Lewin Benjamin (ed.) Genes IX
Подождите немного. Документ загружается.


eral
proteins;
the U4lU6
snRNP
contains two
snRNAs and
several
proteins.
Some
proteins
are common to all
snRNP
particles.
The
snRNPs
recognize
consensus sequences.
Ul snRNA base
pairs
with the 5'
splice site,
U2 snRNA
base
pairs
with
the branch
sequence, and
U5 snRNP
acts at the 5' splice
site. When
U4 releases
U6,
the
U6 snRNAbase
pairs
with U2;
this
may
cre-
ate the catalytic center for
splicing.
An alter-
native
set of snRNPs
provides
analogous
functions for
splicing the
U12-dependent
sub-
class of introns. The
snRNA molecules
may
have
catalytic-like roles in
splicing and
other
processing
reactions.
In
the nucleolus,
two
groups
of snoRNAs
are responsible for
pairing
with rRNAs at
sites
that are
modified;
group
C/D snoRNAs indicate
target sites for methylation,
and
group
ACA
snoRNAs identify
sites where
uridine is con-
verted to
pseudouridine.
Splicing is usually
intramolecular,
but tr ans
-
splicing
(intermolecular
splicing)
occurs in try-
panosomes
and nematodes.
It involves
a
reaction between
a small SL RNA
and the
pre-
mRNA.
The SL RNA resembles
Ul snRNA
and
may
combine the role
of
providing
the exon
and the functions
of UI. In worms
there are
two types of SL RNA:
One is used for
splicing
to the 5'end
of an mRNA, and
the other is used
for
splicing to an internal
site.
Group
II introns
share
with nuclear introns
the use of a lariat as intermediate,
but are able
to
perform
the reaction
as a self-catalyzed
prop-
erty of the
RNA.
These introns
follow the GT-
AG rule, but form a
characteristic secondary
structure that
holds
the reacting
splice sites in
the appropriate apposition.
Yeast IRNA splicing involves
separate endonu-
clease and ligase reactions.
The endonuclease rec-
ognizes the secondary
(or
tertiary)
structure of
the
precursor
and cleaves both
ends of the
intron.
The two half-tRNAs releasedbyloss
of
the intron
can be ligated in the
presence
of ATP.
The termination
capacity
of
RNA
poly-
merase II has not
been characterized,
and 3'
ends of
its transcripts
are
generated
by
cleav-
age.
The
sequence AAUAAA, Iocated I I
to 30
bases upstream
of the cleavage site,
provides
the signal for both cleavage
and
polyadeny-
Iation. An
endonuclease and the
poly(A) poly-
merase
are associated in a complex
with other
factors that confer specificity for
the
AAUAAA
signal.
Transcription is
terminated
when
an
exonuclease, which binds to the 5'end
of
the
nascent RNA chain
created by the cleavage,
catches up
to RNA
polymerase.
Reviews
Kererences
Introduct'on
Reviews
Dreyfuss,
G., I(m, V. N.,
and I(ataoka, N.
(2002).
Messenger-RNA-binding
proteins
and the
messages
they
carry. Nat.
Rev. Mol. Cell Biol.3
t95-205.
Dreyfuss,
G. et
al.
(1993).
hnRNP
proteins
and the
biogenesis of
rrRNA.
Annu. Rev. Biochem.62,
289-J2t
Lewin,
B .
ll97
5l .
Ilnits of transcription
and trans-
lation: sequence components
of
hnRNA and
mRNA. Cell 4,77-93.
Nuctear
Splice
Junctions
Are Short
Sequence:;
Padgefi, R. A.
(1986).
Splicing
of
messenger RNA
precursors.
Annu
Rev.Biochem
55,
Ill9-1150.
Sharp, P. A.
(1987).
Splicing
of
mRNA
precursors.
Science 2)5,
766-771.
Pre-mRNA Spticing
Proceeds through
a
Lariat
Reviews
Sharp,
P. A.
(t
994). Split
genes
and RNA splicing.
Cell
77
,
805-8
t 5.
Weiner,
A.
(1993).
mRNA splicing
and autocat-
alytic introns: distant
cousins
or the
products
of chemical determinism.
Cell
72, 16l-164.
Resea rch
Reed,
R. and
Mani,rtis, T.
(1985).
Intron sequences
involved in
lariat formation
during
pre-mRNA
splicing. Cell
4l, 95-105.
Reed, R.
and
Mani,rtis,
T.
(1986).
A role for exon
sequences and splice-site
proximity
in splice-
site selection.
'iell
46, 681-690.
Zhtang, Y. A., Goklstein,
A. M.,
andWeiner,
A. M.
(
1989) . UACUAAC
is the
preferred
branch
site
for mammalian
mRNA splicing.
Proc. Natl.
Acad.
Sci. USA
56,2752-2756.
snRNAs
Are Required
for
Splicing
Reviews
Guthrie, C.
(1991)
Messenger
RNA
splicing
in
yeast:
clues to
why the
spliceosome
is a
ribonucleoprotein.
Science
25),
157
-163.
Guthrie, C.
and Par.terson,
B.
(1988).
Spliceosomal
snRNAs.
Annu. Rev. Genet.
22, 387-419.
Maniatis,
T.
and
Rt'ed,
R.
(1987).
The role of small
nuclear ribonrLcleoprotein
particles
in
pre-
mRNA splicinl;.
Nature
)25, 67)-678.
Resea rc h
Grabowski,
P.
J.,
St:iler,
S. R., and
Sharp,
P. A.
(
I 985
).
A multicomponent
complex
is
References 707

involved in
the splicing of
messenger
RNA
precursors.
Cell
42,
345-353.
Zhou, 2., Licklider, L.
J., Gygi, S.
P.,
and Reed, R.
(2002).
Comprehensive
proteomic
analysis
of the
human
spliceosome. Nature 419,
I
82-I 85.
@
U1 snRNP Initiates
Spticing
KEVlEW
Brow, D. A.
(2002).
Allosteric cascade
of spiiceo-
some activation. Annu Rev.
Genet.36,
33)-360.
Resea
rch
Abovich,
N. and
Rosbash, M.
(I997).
Cross-intron
bridging
interactions
in the
yeast
commitment
complex are conserved in mammals
Cell 89,
40)-4),2.
Berglund,
J. A., Chua, I(., Abovich,
N., Reed, R.,
and Rosbash, M.
(1997).
The
splicing factor
BBP interacts
specifically with the
pre-mRNA
branchpoint
sequence UACUAAC.
Cell 89,
781-787.
Burgess,
S., Couto, J.
R.,
and Guthrie, C.
(1990).
A
putative
ATP
binding
protein
influences the
fidelity
of branchpoint
recognition
in
yeast
splicing.
Cell 60,
7
O5-7 17.
Parker,
R.,
Siliciano,
P.
G., and Guthrie,
C.
(1987).
Recognition
of the TACTAAC
box during
mRNA
splicing
in
yeast
involves base
pairing
to the
U2-like snRNA. Cell 49, 229-2]9.
Singh, R., Valcdrcel,
J., and Green, M. R.
(1995).
Distinct
binding
specificities and functions of
higher
eukaryotic
polypyrimidine
tract-bind-
ing
proteins.
Science
268,
llT
j-\176.
Wu,
S.,
Romfo,
C. M., Nilsen, T.
W., and Green,
M. R
(1999).
Functional recognition
of the 3'
splice site AG by the
splicing factor U2AF35.
Nature
402,8)2-835.
Zamore, P.
D. and Green, M. R.
(1989).Identifica-
tion,
purification,
and biochemical
characteri-
zation of
U2 small
nuclear
ribonucleoprotein
auxiliary f.actor. Proc.
Natl. Acad.
Sci. USA 86,
924)-9247.
Zhang, D.
and Rosbash, M.
(1999).
Identification
of
eight
proteins
that
cross-link to
pre
-mRNA
in
the
yeast
commitment
complex.
Genes
Dev.
r),
t8t-592.
Zhuang, Y.
and Weiner, A. M.
(1986).
A compen-
satory
base change in
UI snRNA suppresses
a
5
splice site mutation.
Cell 46, 827-B)5.
@
The E
Comptex Can Be Formed
by Intron
Definition
or Exon Definition
Research
Bruzik,
J. P. and Maniatis,
T.
(1995).
Enhancer-
dependent
interaction
between 5'
and 3'
splice
sites in trans- Proc.
Natl Acad
Sci. USA 92,
7056-7059,
5 snRNPs
Form
the Soliceosome
Reviews
Kramer, A.
(I996).
The
structure and function
of
proteins
involved in mammalian pre-
mRNA splicing.
Annu. Rev.
Biochem.65,
367409.
Madhani,
H. D. and Guthrie, C.
(1994).
Dynamic
RNA-RNA interactions in the
spliceosome.
Annu Rev.
Genet
28, l-26.
Resea rch
Lamond, A. I.
(1988).
Spliceosome assembly
involves
the binding and
release
of
U4
small
nuclear ribonucleoprotein.
Proc. Natl. Acad
Sci.
usA 85, 4tt4r5.
Lesser,
C.
F.
and Guthrie, C.
(1993).
Mutations in
U6 snRNA that alter splice site specificity:
implications
for the active site. Science 262,
l
982-r 988.
Madhani, H. D. and Guthrie, C.
(19921.
A novel
base-pairing interaction between
U2 and U6
snRNAs suggests a
mechanism
for
the cat-
alytic activation of the spliceosome.
CellTI,
80 3-8 1 7.
Newman,
A. and
Norman, C.
(1991).
Mutations in
yeast
U5 snRNA alter the
specificity of 5'
splice
site cleavage. Cell 65,
ll5-123.
Sontheimer, E. J. and Sreirz, J. A.
(1993).
The U5
and U6 small nuclear RNAs
as active site com-
ponents
of the spliceosome. Science 262,
1989-r996.
An Atternative
Spticing Apparatus
Uses
Different
snRNPs
Resea
rc h
Burge,
C.
B., Padgett, R. A.,
and Sharp, P. A.
(1998).
Evolutionary
fates and origins
oI Ul2-
type introns. Mol.
Cell
2, 77)-7
85 .
Dietrich,
R. C., Incorvaia, R., and Padgett,
R. A.
'1997 ).
Terminal
intron dinucleotide
sequences do not distinguish
between
U2- and
Ul2-dependent introns. MoL
Cell 1, I
5l-I60.
Tarn, W-Y.
and Steitz, J.
(1996).
A novel
spliceo-
some containing Ul l, Ul2,
and U5 snRNPs
excises a minor class AT-AC intron
in vitro.
CelI
84,801-811.
Spticing
Is
Connected to Export
of
mRNA
Reviews
Dreyfuss,
G., I(m, V.
N.,
and Kataoka,
N.
(2002).
Messenger-RNA-binding
proteins
and the
messages
they carry. Nat Rev.
Mol. Cell BioL
3,
195-205.
Reed,
R. and Hurt, E.
(2OO2l
. A conserved
mRNA
export machinery
coupled to
pre-mRNA
splic-
ing.
Cell 108, 523-5j1.
702
CHAPTER 26
RNA Spticing
and Processing

Resea rc h
I(ataoka,
N.,
Yong,
J., I(im, V.
N, Velazquez, F.,
Perkinson,
R. A.,
Wang,
F,,
and Dreyfuss,
G.
(2000).
Pre-mRNA
splicing imprints mRNA
in
the
nucleus
with a novel RNA-binding
protein
that
persists
in the
cytoplasm. Mol
Cell
6,
673-682.
Le Hir, H., Gatfield, D.,
Izaurralde, E.,
and
Moore,
M.
J.
(2001).
The
exon-exon
junction
com-
plex provides
a binding
platform
for factors
involved in mRNA
export and nonsense-
mediated
mRNA decav. EMBO
J.20.
49874997.
Le Hir, H., Izaurralde, E.,
Maquat, L. E.,
and
Moore, M.
J.
(2000).
The spliceosome
deposits
multiple
proteins
20-24 nucleotides
upstream
of
mRNA
exon-exon
iunctions.
EMBO J.19.
6860-6869.
Luo, M.
J. and
Reed,
R.
(1999).
Splicing
is required
for rapid and
efficient mRNA
export
in meta-
zoans. Proc Natl. Acad.
Sci. USA 96,
t49)7-14942.
Luo, M. L., Zhou,2., Magm,I(.,
Christoforides,
C.,
Rappsilber,
J.,
Mann,
M., and Reed, R.
(2001).
Pre-mRNA splicing and mRNA
export linked
by direct interactions
between UAP56 and
Aly. Nature 4l), 644=647
Reichert,
V. L., Le Hir, H.,
Jurica, M. S., and Moore,
M. J.
(2002).
5'exon interactions
within the
human
spliceosome establish
a
framework for
exon
junction
complex
structure and assem-
bly.
Genes
Dev. 1
6,
277
8-27 9 1.
Rodrigues,
J.
P., Rode, M.,
Gatfield, D., Blencowe,
B., Blencowe, M.,
and Izaurralde, E.
(2001).
REF
proteins
mediate the export
of spliced
and unspliced mRNAs from
the
nucleus.
Proc.
Natl. Acad. Sci USA 98, t010-1035.
Strasser,
I(.
and
Hurt,
E.
(2001).
Splicing
factor
Sub2p
is required
for nuclear mRNA
export
through its interaction wrthYralp.
Nature
4t),648-652.
Zhou,2., Luo, M. J.,
Straesser, I(., I(atahira, J.,
Hurt, E.,
and
Reed,
R.
(2000).
The
protein
AIy
links
pre-messenger-RNA
splicing to
nuclear
export in metazoans. Nature 407,
4Ol-405.
Group II Introns Autosptice
via
Lariat
Formation
Michel, F.
and
Ferat,
J.-L.
(1995).
Structure and
activities of
group
II introns. Annu Rev.
Biochem. 64, 4)5-461.
Alternative SpLicing Invotves Differential.
Use
of Solice
Junctions
Review
Green, M.
R.
(
I
99
I
).
Biochemical mechanisms of
constitutive and regulated
pre-mRNA
splicing.
Annu. Rev. Cell Biol.7. 559-599.
Review
Resea rc h
Handa. N.. Nureki, O.,
I(urimoto,
I(., ICm,
I.,
Sakamoto, H., Shimura,
Y.,
Muto, Y., and
Yokoyama, S.
(1999).
Structural
basis for
recognition of the
tra mRNA
precursor
by the
Sex-lethal
protein.
Nature )98,
579-585,
Lynch, I(. W. and
X{aniatis, T.
(1996).
Assembly of
specific
SR
prc,tein
complexes
on
distinct
reg-
ulatory elements of the
Drosophila
doublesex
splicing enhancer.
Genes
Dev.10,
2089-2101.
Sun,
Q.,
Mayeda,
l\., Hampson,
R. K.,
I(rainer,
A. R.,
and
Rottman,
F. M.
(19931. General
splicing
factor SF2/ASF
promotes
alternative
splicing by binding
to an exonic
splicing
enhancer. Genes
Dev.
7, 2598-2608.
Tian, M. and Mani,rtis,T.
(19931.
A splicing
enhancer
complex
controls alternative
splic-
ing of doubles:x
pre-mRNA.
Cell74,
105-l14.
Wu, J. Y. and Maniatis
,
T.
(1993).
Specific
interac-
tions between
proteins implicated
in splice site
selection and
leguiated
alternative splicing.
Cell75,l06l-t070.
trans-Sp[icing
Reactions
Use Smat[
RNAs
Review
Nilsen,
T.
(1993).
l?ans-splicing
of nematodepre'
mRNA. Annu.
Rev. Immunol.
47, 413-440.
Resea rc h
Blumenthal, T., Evans,
D.,
Link, C. D., Guffanti,
A.,
Lawson, D., Thierry-Mieg,
J.,
Thierry-Mieg,
D.,
Chiu, W.
L., Duke,
I(., ICralY,
M., and
I(m,
S. K.
(2002).
A
global analysis of C
elegans
operons.
Natu"e 417, 85
l-854.
Hannon, G. J. et al.
(1990).
trans-splictng
of
nema-
tode
pre-mRN A in vitro Cell
61,
1247-1255.
Huang, X. Y. and Hirsh,
D.
(1989).
Asecondtrans-
spliced
RNA le ader sequence
in the nematode
C elegans.
Proc Natl.
Acad Sci. USA
86,
8640-8644.
Krause, M. and Hilsh,
D.
(1987).
Atrans-spliced
leader sequen,:e
on actin
mRNA
tn
C
elegans.
Cell
49
,
7 5)-7
'51
.
Murphy, W. J., Watkins,
K. P., and
Agabian, N.
(
l936). Identilication
of
a novel
Y branch
structure as
art intermediate
in trypanosome
mRNA
proce
s r;ing
:
evidence
for tr an s
-
splicing.
Cell 47, 517-525.
Sutton,
R. and Boc'throyd,
J. C.
(1986). Evidence
f.or trans-splicing
in trypanosomes.
Cell
47,
527-535.
The Spticing
Endonuctease
Recognizes
tRNA
Resea
rch
Baldi, I. M. et al.
|
9921 . Participation
of the
intron
in the
reaction catalyzed
by the
Xenopur IRNA
splicing endorruclease
. Science
255,
t404-1408.
References
703

Diener,
J. L. and Moore, P. B.
(1998).
Solution
structure of
a substrate for the archaeal
pre-
tRNA splicing endonucleases:
the bulge-helix-
bulge motif. Mol.
Cell
l,
88)-894.
Di Nicola
Negri, E., Fabbri,
S., Bufardeci, E., Baldi,
M. L, Mattoccia,
E., and Tocchini-Valentini,
G.
P.
(1997).
The eucaryal
IRNA splicing
endonuclease recognizes
a tripartite set of
RNA elements.
Cell 89. 859-866.
Kleman-Leyer,
I(.,
Armbruster, D. W.,
and
Daniels,
C. J.
(2000).
Properties
of H.
volcanii
IRNA
intron endonuclease
reveal a relationship
between
the archaeal and
eucaryal IRNA
intron
processing
systems. Cell 89,8)9-847.
Lykke-Andersen,
J. and Garrett, R. A.
(1997).
RNA-protein
interactions of
an archaeal
homotetrameric
splicing endoribonuclease
with an
exceptional evolutionary history.
EMBO
J
16,6290-6300.
Mattoccia,
E. et al.
(
1988) .
Site selection by the
tRNA splicing
endonuclease of. X.laevis
Cell
55, 7jt-738.
Reyes,
V. M.
and Abelson, J.
(1988).
Substrate
recognition
and splice site
determination in
yeast
IRNA splicing.
Cell 55,719-7)0.
Trotta,
C. R., Miao, F., Arn, E.
A., Stevens,
S. W,
Ho, C. K., Rauhut,
R., and Abelson,
J. N.
(1997).
The
yeast
tRNA splicing
endonucle-
ase: a tetrameric
enzyme
with two active site
subunits homologous
to the archaeal
IRNA
endonucleases.
Cell 89,
849-858.
The
Unfolded Protein
Response Is
Retated
to IRNA Spticing
Gonzalez,
T. N.,
Sidrauski, C., Dorfler,
S
,
and Wal-
ter, P.
(1999).
Mechanism
of non-spliceosomal
mRNA
splicing in
the unfolded
protein
response pathway.
EMBO J.18,
)l19-382.
Sidrauski,
C., Cox, J. S., and
Walter, P.
(1996).
IRNA ligase is required
for regulated
mRNA
splicing
in the unfolded
protein
response.
Cel/
87, 405-4t).
Sidrauski,
C. and Walter, P.
(1997).
The
transmem-
brane kinase Irelp
is a site-specific
endonucie-
ase that
initiates mRNA
splicing in
the
unfolded
protein
response.
Cell
90,
I 03 l-r
039.
@
The
3' Ends of mRNAs
Are Generated
by
Cteavage and Potyadenylation
Review
Wahle,
E.
and I(eller, W.
(1992).
The
biochemistry
of l'-end
cleavage and
polyadenylation
of
messenger
RNA
precursors.
Annu. Rev.
Biochem
61,419-440.
Resea rch
Bouvet,
P.,
Omilli, F., Arlot-Bonnemains,
Y.,
Legagneux,
V.,
Roghi,
C., Bassez,
T., and
Osborne,
H. B.
(1994).
The deadenylation
CHAPTER
26
RNA Spticing
and Processing
@
Researc h
conferred by the 3'untranslated
region of a
developmentally
controlled
mRNA
in Xenopus
embryos is switched to
polyadenylation
by
deletion of a short sequence
element. Mol. Cell
Biol. 14, 1893-1900.
Conway,
L.
andWickens, M.
(1985).
Asequence
downstream of AAUAAA is required
for for-
mation
of SV40
late mRNA
3'termini in frog
oocytes. Proc. Natl. Acad. Sci. USA
82,
j949-3953.
Fox,
C.
A.,
Sheets,
M.
D., and Wickens, M. P.
(I989).
Poly(A) addition during maturation
of
frog oocytes: distinct nuclear
and cytoplasmic
activities and regulation
by the sequence
UUUUUAU. Genes Dey. ).2151-2162.
Gil, A. and Proudfoot,
N.
(1987).
Position-
dependent sequence elements
downstream
of AAUAAA are required for
efficient rabbit
B-globin
mRNA
3'end
formation.
Cell
49,
)99406.
Karner, C. G., Wormington, M., Muckenthaler,
M.,
Schneider,
S.,
Dehlin, E.,
and Wahle, E.
(1998).
The deadenylating nuclease (DAN)
is
involved in
poly(A)
tail removal during
the
meiotic maturation
of Xenopus oocytes.
EMBO
J.
17,
5427-54)7 .
ICm, M., I(rogan,
N. J., Vasiljeva, L., Rando,
O. J.,
Nedea,
E.,
Greenblatt, J.
F.,
and Buratowski,
S.
(2OO4l.
The
yeast
Ratl
exonuclease
promotes
transcription
termination by RNA
polymerase
II. Nature 432,
517-522.
McGrew,
L. L., Dworkin-Rastl, E., Dworkin,
M. B.,
and Richter,
J.
D.
(1989).
Poly(A)
elongation
during Xenopus
oocyte
maturation
is required
for translational
recruitment and is mediated
by a
short sequence element. Genes Dev.
),
80 3-8
l
5.
Takagaki, Y., Ryner, L.
C., and Manley,
J. L.
(1988).
Separation and characterization
of a
poly(A)
polymerase
and a cleavage/specificity factor
required for
pre-mRNA polyadenylation.
Cell
52,73t-742.
Yoeltz,
G.
I(.
and Sreitz, J. A.
(I998).
AUUUA
sequences direct mRNA
deadenylation
uncou-
pled
from
decay during Xenopus
early develop-
melrl
Mol. Cell Biol. 18.7537-7545.
Cteavage of
the 3'
End
of Histone mRNA
May
Require a Sma[[
RNA
Review
Birnstiel,
M. L.
(
I 985
).
Transcription
termination
and 3'processing:
the end is in
site. Cell 41,
349-359.
Re
sea
rc h
Bond,
U. M., Yario, T. A.,
and Steirz,
J. A.
(1991).
Multiple
processing-defective
mutations
in a
mammalian histone
pre-mRNA
are sup-
pressed
by compensatory
changes in
U7 RNA
bolh in vitro
and in vitro.
Genes Dev. 5
,
t709-t722.
704

Dominski,2., Erkmann,
J. A., Greenland,
J. A.,
and
Marzluff,
W. F.
(2001).
Mutations in
the
RNA binding domain
of stem-loop
binding
protein
define separable requirements
for
RNA
binding and for histone
pre-mRNA pro-
cessing. Mol. CelI. Biol. 21,
2008-2017 .
Galli,
G.
et al.
(1983).
Biochemical
complementa-
tion with
RNA
in lhe Xenopus
oocyte: a small
RNA is required
for the
generation
of 3'his-
tone
mRNA
termini. Cell
)4, 821-828.
Mowry, I(.
L. and Steitz,
J.
A.
(1987).
Identifica-
tion of
the
human
U7 snRNP
as one of several
factors involved in
the 3'end maturation
of
histone
premessenger
RNAs.
Science
2)8,
t682-t687.
Wang,
Z. F.,
Whitfield, M. L., Ingledue,
T. C.,
Dominski, 2., and Marzluff, W.
F.
(1996).
The
protein
that binds
the 3' end of histone
mRNA: a novel RNA-binding protein
required
for histone
pre-mRNA processing.
Genes Dev.
r0,3028-3040.
Production
of rRNA
Requires Cteavage
Events
Venema, J. and Tollervey, D.
(1999).
Ribosome
synthesis in S. cerevisiae. Annu.
Rev. Genet. 33,
26t-3tt.
l@
Smal.L RNAs
Are
Required for rRNA
Processi ng
Resea rc h
Balakin, A.
G., Smith,
L.,
and Fournier, M. J.
(1996\.
The RNA world
of the nucleolus: two
major families of small RNAs
defined by dif-
ferent box elements
with related functions.
Cell 86, 82)-814.
Bousquet-Antonelli, C., Henry,
Y., G'elugne, J.
P.,
Caizergues-Ferrer,
M., and
Kiss, T.
(1997). A
small nucleolar RNP
protein
is
required for
pseudouridylation
of eukaryotic
ribosomal
RNAs. EMBO J. 16, 477 0-477
6.
Ganot, P., Bortolin,
M. L., and
I{ss, T.
(I997).
Site-
specific
pseudouridine formation
in
preriboso-
mal RNA is
guided
by small
nucleolar
RNAs
Cell 89,
799-81)9 .
Ganot,
P.,
Caizergues-Ferrer,
M., and
Kiss, T.
(19971
.
The family of box
ACA small
nucleo-
lar RNAs is defined
by an evolutionarily
con-
served secondary
structure
and ubiquitous
sequence elerrrents
essential
for RNA
accumu-
Iation. Genes
Dev. ll,94l-956.
I(ass,
S. et
al.
(1990).
The U3 small
nucleolar
ribonucleoprotein
functions
in
the
first step
of
preribosomal
RNA
processing.
Cell 60,
897-908,
ICss-Laszlo, Z.,Henry,
Y., and
Kiss, T.
(1998).
Sequence and structural
elements
of
methyla-
tion
guide
snoRNAs
essential
for site-specific
ribose methylation
of
pre-rRNA. EMBO J. I7,
797-807.
I(iss-Laszlo, Z.
et
al.
(19961
. Site-specific
ribose
methylation oI
preribosomal
RNA:
a novel
function for srnall
nucleolar
RNAs. Cel/
85,
1077-t068.
Ni, J.,
Tien, A. L., and
Fournier,
M. J.
(1997).
Small
nucleolar RNAs
direct site-specific
syn-
thesis of
pseuctouridine in rRNA. Cell
89,
565-57 i.
Review
References
705
Catalytic
RNA
I
CHAPTER
OUTLINE
Introduction
Group I Introns
Undertake
SeLf-SpLicing
by
Transesterification
o
The
only
factors
required
for
autospticing in vitro
by
group
I
introns
are a monovatent
cation,
a divalent cation,
and a
guanine
nucteotjde.
o
SpLicing
occurs by
two transesterifications,
without requir-
ing input
of energy.
r
The 3'-0H
end
of the
guanine
cofactor
attacks the 5'end of
the intron in
the first
transesterification-
r
The
3'-0H end
generated
at the end of the first
exon attacks
the
junction
between
the intron
and second exon in
the
second
transesterifi
cation.
.
The intron
is reteased
as a [inear motecu[e
that circutarizes
when its
3'-0H
terminus
attacks a
bond at one oftwo inter-
naI
oositions.
.
The
G414-A16 internal
bond of
the
intron
can
also be
attacked by
other nucteotides
in
a trans-splicing reaction.
Group I Introns
Form
a Characteristic
Secondary Structure
r
Group I introns
form
a secondary
structure with nine
duplex
reglons.
r
The
cores
of regions
P3, P4, P6,
and P7 have
catalytic
activity.
.
Regions
P4 and P7
are
both
formed
by
pairing
between con-
served
consensus
sequences.
e
A
sequence
adjacent
to P7
base
pairs
with
the sequence
that
contains
the reactive
G.
Ribozymes Have
Various
Catatytic Activities
o
By
changing
the substrate
binding-site
of
a
group
I intron,
it is
possible
to introduce
alternative
sequences
that
jnter-
act with
the reactive
G.
o
The reactions
follow
ctassical enzvme
kinetics with
a low
catatytic rate.
o
Reactions
using 2'-0H
bonds
coutd have
been the basjs for
evolving
the
originaI
catalytic
activjties in RNA.
Some
Group I Introns
Code for Endonucleases
That
Sponsor
Mobil.ity
.
Mobite
introns
are abte
to insert
themselves into new
sites.
o
Mobi[e
group
I introns
code for
an endonuctease
that makes
a double-strand
break at
a target site.
o
The
intron
transposes
into
the site
of the doubte-strand
break by a DNA-mediated
repticative
mechanism.
Group Ii
Introns
May
Code for Muttifunction
Proteins
o
Group II
introns
can
autosplice in vitro,
but
are usualty
assisted
by
protein
activities
coded within
the intron.
706
.
A singte coding frame specifies a
protein
with reverse
tran-
scriptase activity, maturase activity, DNA-binding
motif,
and
a
DNA
endonuclease.
.
The reverse
transcriptase
generates
a DNA
copy of the RNA
sequence that transposes by a retroposon-tike
mechanism.
o
The
endonuclease cleaves
target
DNA
to a[[ow insertion
of
the transposon at a new site.
Some Autospticing Introns
Require Maturases
o
Autospticing
introns may require maturase
activities
encoded within the intron to assist fotding
into
the active
catatytic structure.
The Catatytic Activity
of RNAase P Is Due
to RNA
r
Ribonuctease P is
a ribonucteoprotein in
which the RNA has
catalytic
activity.
Viroids Have Catatytic Activity
.
Viroids
and virusoids form a hammerhead
structure
that has
a
setf-cleaving activity.
o
Simitar structures can be
generated
by
pairing
a substrate
strand
that
is
cteaved by an enzyme
strand.
.
When an
enzyme strand is introduced into
a cett, it
can
pair
with a substrate
strand target that is
then cteaved.
RNA Editing
0ccurs at
Individua[
Bases
o
Apo[ipoprotein-B
and
glutamate
receptors
have site-specific
deaminations catatyzed
by cytidine and adenosine
deami-
nases
that change
the coding sequence.
RNA Editing
Can Be Directed
by Guide RNAs
.
Extensive
RNA editing in
trypanosome mitochondria
occurs
by
insertions
or deletions
of uridine.
o
The substrate
RNA base
pairs
with
a
guide
RNA
on both
sides of the region
to be edited.
r
The
guide
RNA
provides
the
temptate for addition
(or
Less
often, deletion)
of uridines.
r
Editing is
catalyzed
by a complex of
endonuclease,
terminal
uridyttransferase
activity,
and RNA ligase.
Protein
Spticing Is Autocatalytic
o
An intein
has
the abitity to catatyze its
own removal
from
a
protein
in such
a way that the flanking
exteins
are
connected.
r
Protein
spticing is catatyzed
by the intein.
o
Most inteins have
two independent
activities:
protein
splic-
ing
and a homing
endonuclease.
Summary
@
169
slua^l
a6p^pall
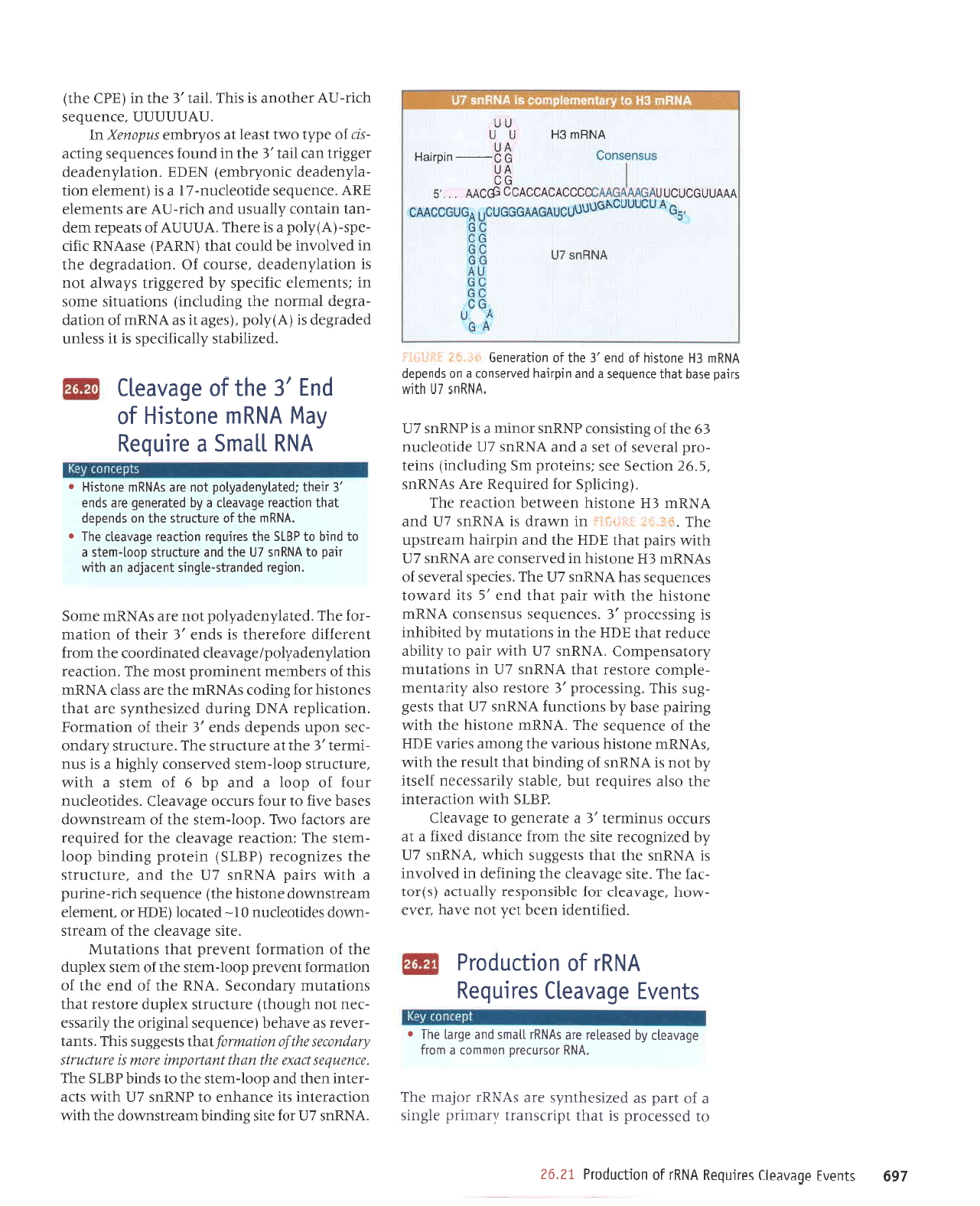
serrnbau
vNUr
Jo
uoqlnpold
lz'gz
ol
pJsseJoJd
sr
1eq] lduJsueJt
Lrerurrd
a18urs
e
1o
ged
se
pezrseqluz(s
are
syNUr roferu aq1
'vNU
roslnJalo uourrrol e uroj]
a0eneep fiq
pasealar
ele
sVNUI
llputs
pue
a6te1 aq1
e
vNUr
Jo
uor+lnpold
'pJlJrluepr
uaaq
la,{.
lou
e^eq
'JJAJ
-uoq
'aBeneJIJ
ro;
alqrsuodsar Lllenpe
(s)ro1
-JPJ
aqJ
'Jlls
eSe^eelr
aql Sururyap ur
pJAIOAUI
sI
yNUus
Jqt
leqt
stseSSns
qrlqM
'vN5us
4n
z{.q
pazruSorJJ
etrs
eq1 uolJ
Jf,uelsrp
pJXrJ
p
tp
sJnJJo snururJJl,€
p
JleJeue8 o1 a8eneal3
agfs
qllM
uoIllPJJluI
Jql
oslp sarrnbar
tnq
'Jlqpls
.dpressarau
11asl1
z(q
tou
sr
VNUUs
yo
Surpurq
leqt tlnsar
rqt
qlr^{
'syN5ru
Juolsrq
snorJEA aql Suoure sJrre^
A(H
aqt
Jo
aJurnbas
aq1
'VNUr.u
Juotsrq eql
qllm
SurrrBd
aseq Lq suorlJunl
VNUus /n
tpqt
stse8
-3ns
srql
'Surssarord
,€
eJolseJ osle dlrreluaur
-aldruor
eJotsar
tpql
vNuus
Zn
ur suortptnu
,{rolesuadruol
'VNUus
Zn
qtlM
rred ot,{tq1qe
JJnpJr
teql
g(H
rqt
ur suorlptnru
,{q
pJtlqlqul
sr Surssarord
,g
'saruanbas
snsuJsuoJ
VNUtu
Juolsrq aql
qll,lr
rrBd
teql
puJ
,S
slr
pJplnrol
saruanbas
seq
vN5us 4n
aqJ
'sar;ads
IeJJAes Jo
svNuru
€H
Juolsrq
ur
pJ^resuoJ
aJe
vNuus /o
qlltzr
srted
lPql
iI(H
aqt
pue
urdrreq ruearlsdn
e{I
',,r'rr
r
rtr.iiriir
I
UI UMptp SI
YNUUS
/n
pup
vNIUru €H
Juolsrq
uJJMlJq
uortf,eJJ
aqJ
'(3uo11dg
ro;
parrnbag
rrv sVNUus
'S'92
uorlJJS
aas
isuratord
ug Surpnltur)
sural
-ord
leranas
Jo
tas
e
pue
vNuus /|l
aprtoJlJnu
€9
Jql
Jo
Surlsrsuor
dN5us
Jourur e sr
dNUus /n
'vNUus
/n
qlm
slred
aspq
1eq1
aruanbes
e
pue
urdtLeq
pa^lasuol
e uo
spuadep
vNUtx €H
auolsrq
J0
puo
/t
aql
Jo
uorlPreue9
r.;
',.;
r:!{i1-.l,iri
'vNuus
/nroJ
Jlrs durpurqueJrlsuMop Jql
qlrM
uorlJeJalur slr aJupque ol
dNuus zn
qlIM
sltp
-rJlur
uJql
pue
dool-ruals eq1 01 spurq
dg'IS
eqJ
'atuanbas
paxa
aqi uaLfi
ruuuldu,fi
alu,Lt
st alnpnus
[npuons atlllo uo4awoltpqt s1se33ns srqJ
'stue]
-ralJr
se Jleqaq
(aruanbas
put3rro
aqt ^lupsse
-JJu
lou
q8noqt)
aJnDnJls xaldnp Jrolsar
ler{l
suortptnur
.drepuora5
'VNU
aql
Jo
pue
eq1
Jo
uoppruJoJ
luanard
dool-rua1s eql
Jo
rrrels xaldnp
Jql
JO
uorleuroJ
luarra.rd leql
suorlelnw
'a1rs
a8errealJ Jqr
Jo
urpJrls
-rrmop
sJprloelJnu
0I-
pJlprol (gOg
ro
'luJruelr
rueeJtsumop auolslq aqt)
aruenbas
qlr-aur,rnd
e
qlrm
srred
yggus
4n
eql
pup
'eJnlJnJls
aqt sazruSorar
(4919)
uralord Surpurq dool
-tuals
aqJ
:uorlJeeJ
a8erreap eqt
roJ
palnbar
eJp sJolJeJ o.141
'doo1-urals
aql
Jo
upeJlsuMop
sJspq JArJ ot
rnoJ
srnJJo
a8e,{ea13
'sJprtoJllnu
rnol
Jo
dool e
pue
dq
9
Jo
ruals P
qlrM
'aJntJnJls
dool-rua1s
pe^resuo)
.dp8tq
p
sr snu
-rrurJt,€
Jqt
tp
eJnDnJls JqJ
'JJnlJnJls
,{repuo
-ras
uodn spuadap
spuJ
,€
JrJr{l
Jo
uorteruJo{
'uorlerrldar
y1qq
Suunp
pazrsaqluz(s
erp
lpql
sJuolsrq JoJ Surpor svNgru
aqt Jrp ssep
VNUrU
slql
Jo
sraqruJru
luauluord
tsoru
aqJ
'uorlJeJJ
uope1.{uapedlodTa8errealJ
pateurpJoo)
Jqt rrroJJ
lueJeJJrp
aJoJaraql
sr spuJ
,€
Jreql
Jo
uorlPu
-roJ
eql
'patelduape,(1od
tou
Jrp sVNUru JruoS
'uorbar
pepuerls-e16uts
luarelpe
up
qlrm
rred
o1
y11Xus
/n
eql
pup
alnllnlls dool-ue1s e
ol
purq
ol
d€ls
aq] sarrnbar uolpeor abenealr aq1
o
'VNUur
eql
Jo
alnllnlls eq1 uo spuadep
leql
uoqlear
a6eneelr e r\q
paleraueb
arp spuo
,€
rraql
:palelAuapeAlod
1ou
are s!fl!ur ouolstll
o
vNU
llPLus
e arrnbau
,teW
VruUu
auolsr.H
Jo
qua^l
a6enea'13
sarrnbau
wvnn9cncn
ccScvcvccvcc
scw
.
,9
9C
VN
e C-
urdlteH
VN
VNUU EH N N
nn
pul
,t
aql
Jo
e6enea';3
'pazrrqpts
.{gerryrads sl
tr
ssepn
paper8ap
sr
(y)z(1od
'(sa8e
1r
se
VNUIU
Jo
uortep
-er8ap
Ieurou
aql Supnlrur)
suortenlrs auos
ur
islueruelJ
rrynads
Lq
paraSStrl
sziBmlB
lou
sr
uorlelz(uapPJp
'Jsrnol
Jo
'uorleperSap
aqt
ur
pa^Io^u
eq
plnoJ
]Eql
(Nuvd)
JseYNu rrJrr
-ads-
(y)L1od
p
sr rrrql
'vnnnv
Jo
sleadar ruap
-uel
ureluoJ dllensn
pue qJIJ-nV
JJe sluJruele
Egy'aruanbas
eprtoJpnu-4I
p
sl
(tuaruala
uoq
-e1.{uapeap
rruo,{rqrua)
Nfqg'uor1e1z(uapeap
,ra33r:1 upJ
Iret,€
eqt ut
punoJ
saruanbas SutpB
-rr;o
ad,{t oml
tspJl 1e
sozhqura
sndouay u1
-
'nYnnnnn
'aJuenDas
qrrr-nv
reqloup sr slqJ
'llpt
,€
aql
q
(saf
aqt)
lpql
os
tosrnrard
eql 01
purq (suralord.raqlo
.{lqrssod
pue)
suralord
leurosoqrr'Surssarord
ygg.r
rr1o,{.re>lne
pue
rr1o,{relord
qloq
uI
'sn)olutJ
JelDIl
-red
aqt
Io
turluo)
Jqt uo
puadap
snnpord aql
aseJIJJ o1
parmbar
suortJeal
Surssarord aql snqJ
'pue
,€
Jql
pue
aruanbas
SE
Jql uJJMlJq
tue
-sard
aq
lou,{eru.ro
z(eru saua8
VNUI
Ipuoltrppv
'uor8ar
srql ur saua8
VNUI
oMl ureluoJ ooluJJ
JJqlo
rqt
pue
/sJ)uJnbas
y51gr
S€Z
pup
59I
eql ueJMlJq aua8
y551
Juo uIPJuoJ rJol UJJ
rnoJ
lJruouJ8
aqt
punore pasradsrp
are suorado
UJJ uJ^es ertrl
'un'a
uI
'svNul
oMl ro
auo
pue
yNUJ
SS
Jql sureluoJ oslp
rosJnJJrd aqt
teqt
sMoqs
Fr'{,'{,iii l;*:ji.i_i:{
'saruanbas
asaqt uaaulaq 3ur
-ssarord
ou sr rrJql
'sr
lpql
'VNdr
(gE7)
a8rel
er{t
Jo
puJ
,E
aql srurol
VNUr
Sg'E
o1 Surpuods
-JJJoJ
aruanbas JqJ
'erJJDeq
ur rosrntard aql
Jo
uollpzlue8ro aqt ur J)uJreJlrp
p
sr JrJqI
('r{puapuadapur
paqrrJsueJl
sr
tnq
'lrun
uorl
-dtnsuBrl
rolBru qJee qlr,rvr pelerJosse
sr aua8
ss
e
']sea,{
Jo
ese) eql
q)
'syNur
roleur
Jql
loJ
saua8 aqt ruorl aleredJs
JJe
tnq'pJJelsnp
eJe
saua8
5g
aqt
'praua8
uI
'III
Jserrrx,{1od
ygg
Lq
saua8 aleredJs uoJJ
pJqrrtsueJl
sl
VNU SE
'(J)uJnDJS
luPlS
-uo)
ureturpw
VNUJ
JoJ sJUJC
paleadag
aq1
'6'9
uoIDaS aas) sleadar uapupt
se
paztue8ro
are sardor eqJ
'sVNUr
Jqt JoJ
lrun
uorldrrrs
-ueJl
eql
Jo
satdor
aldqlnru sLerr,rp JJe JJJqJ
'sJIn)elolu
eJnlelu Jql eleJJuJS
ol
pasn
aq uer sa8e
-^eelJ
Jo
suorlPurqruoJ
tuJJeJJrp leq]
pue
luep
-unpeJ
aJp sarlrnrtJe rrJqt
lpql
s1sa33ns
qJIqM
'Surssarord
tua.Lard
lou
op Lllensn
saru.{zua
IenpI^Ipur
ur suortelnyX
'(sarlrnglry
a1dr1
-lnw
sJ^lo^u1 uorleper8aq
VNUIU
'€
I'4
uorDes
aas) uoqeperSap
y51gru
ur salednrlred
osp
teqr
saspalJnuoxJ
IeJJAas ;o
,4.1qurasse
ue sr
q)rqM
'eruosoxa
aqt Surpnpul
'VNUJ
Surssatord ur
paterrldrur
uJeq eneq sJspJltnuoqrr
z(ue141
'
uoID ee J Sutruurr.rl,S-,{,
e u(q
p
a,rzrolloy aB err e ap
,(q
paleraua8
are spua
,€
Jql
Jo
lsoyrg
'1uana
a8e
-AeJIJ
p
z(q,{.1narp paleraua8
JJe spuJ,S aql
Jo
lsow
'sa]0fue>lna
IIe
uI
pJAIoAur
eJe suorpeal rel
-runs
,{.11errseq
tnq
'sluala
Jo
JapJo eql
ur suort
-erJen
eq ueJ JrJqI
'lsea^d
ur de,uqled
leraua8
aqt smoqs
lf
',ii,l
.iJil*i:j
'suorlf,eer
Sunuurrrt
puB
sluana a8erreap
Jo
uorleurquor
e
dq
rosrnlard
eql uorJ
paSPJIJJ
are
svNuJ arnlpu
aql
'(tsea,{
ul
SE€)
rrllerus
s1
1r
satorfuelna
JJMoI uI
'vNu
s57
sP Jter
uorlelueurpJs
slr roJ
pJrueu
sr rosrnrard aql
'salodre>1na
raq8rq
uI
'sVNUr
S8Z
pue
'S8'S 'SgI
JqtJo saruanbas
aql surpluoJ
rosrnrard JqJ
'spnpord
arnleru
aql aleraua8
6urssa:or6
pue
6utrqd5
VNU
9Z
UljdVHl
'aprs
.laqlra uo slnr
fiq
1du:s
-up.ll
oql uor1
paspalal
aq
lsnur lrnpord
VNU
qrpl
'pasn
ele (1) stoleututal
pue
(6)
sralouord
qrrqrvr
uo
puadap
sldursuetl aq1lo
sq16ua1
lrpxo
aql'VNU1
pup
VNUI
qloq
loJ saueb
ureluor
qe'l
ur suorado
u/r aql
ijL'1ig
j#ilTl:1
'ldursuerl
rteLuud
p
uo.U s1uala 6uruuul
pue
ebenealr
Aq
palerauab
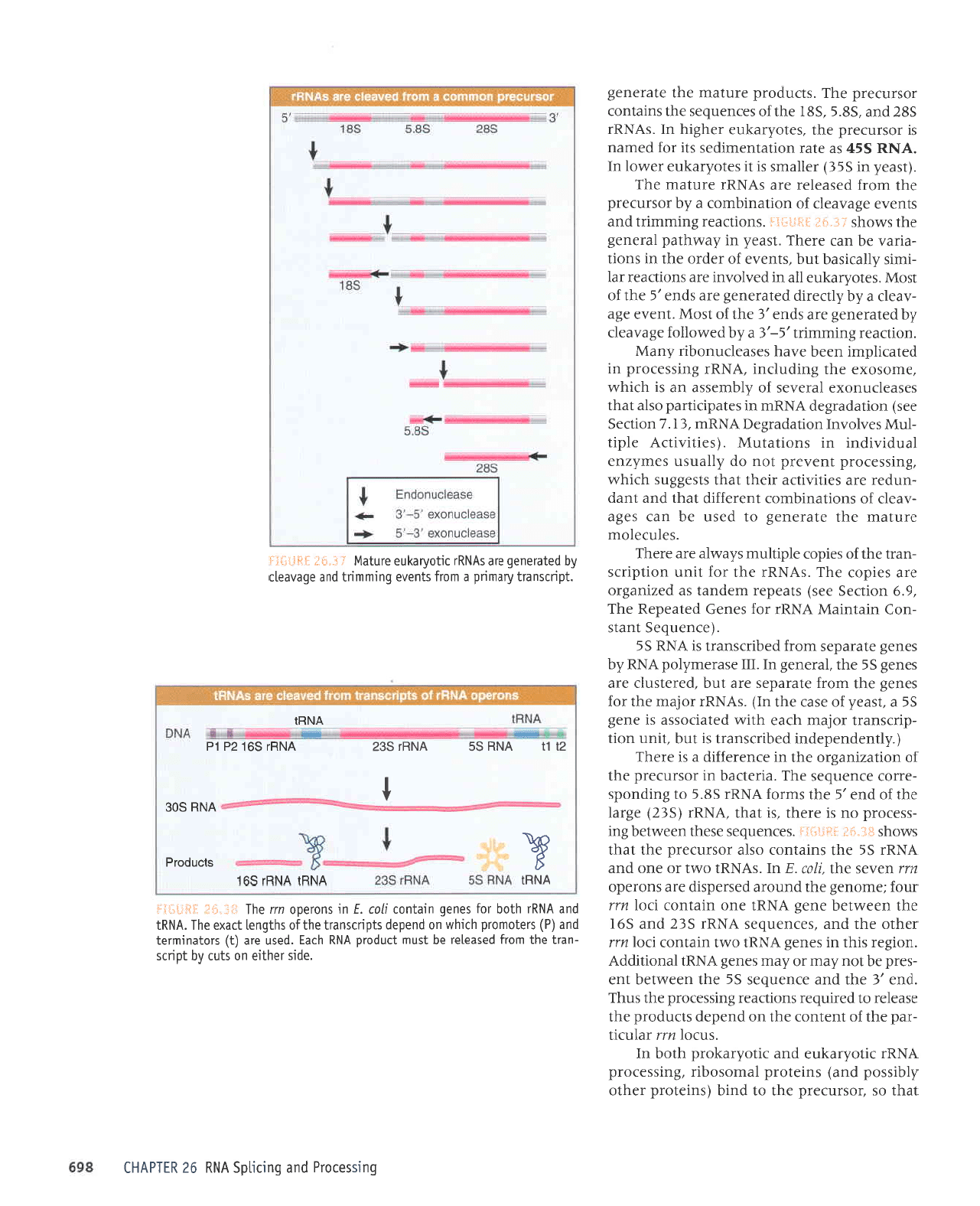
ere sVNUr rrlo&e4na olnlpw
.i i'tI ;igtlt.:t.*
VNUI
I
&
VNU} VNHI 59I
slcnpord
VNU
SOE
I
ZI [I
VNH S9 VNUJ SEZ
vNHr s9!
zd td
VNUT
reopens
the
primary
circle
by reacting
with the
G4r4-A16
bond.
The
UUU
(which
resembles
the
3' end of the l5-mer
released
by the
primary
ryclization)
becomes the 5'end
of the linearmol-
ecule that is formed. This
is an intefinolerularreac-
tion, and thus demonstrates
the ability
to connect
together two
different RNA molecules.
This series of reactions
demonstrates
vividly
that
the autocatalytic activity
reflects a
gener-
alized ability
of the RNA molecule
to form an
active center that can
bind
guanine
cofactors,
recognize oligonucleotides,
and
bring together
the
reacting
groups
in a
conformation that
allows bonds to
be broken and rejoined.
Other
group
I introns
have not been investigated
in as
much detail astlrre Tetrahymenaintron,
but their
properties
are
generally
similar.
The autosplicing
reaction is
an
intrinsic
property
of RNA invitro,
but to what
degree are
proteins
involved
invivo?
Some indications for
the involvement
of
proteins
are
provided
by
mitochondrial systems,
where splicing of
group
I introns requires
lhe trans-acting
products
of
other
genes.
One striking
case is
presented
by
t}re
qttl8
mutant of Neurospora
crassa, which is
defective in splicing
several mitochondrial
group
I introns. The
product
of this
gene
turns out to
be the mitochondrial
tyrosyl-IRNA synthetase!
This is explained
by the fact that the intron
can
take up a tRNA-like
tertiary structure
that is
stabilized by the synthetase
and which
promotes
the
catalytic
reaction.
This relationship
between the
synthetase
and splicing
is
consistent with
the idea that splic-
ing originated as an RNA-mediated
reaction.
subsequently assisted
by
RNA-binding proteins
that originally
had
other functions. The
in vitro
self
-splicing
ability may represent
the basic bio-
chemical
interaction.
The RNA
structure cre-
ates the active site,
but
is
able to function
efficiently in vivo
only when assisted by a
pro-
tein
complex.
@
Group I Introns Form
a Characteristic
Secondary
Structure
Group
I introns form
a secondary structure with
nine
duplex
regions.
The cores of
regions
P3, P4, P6,
and
P7 have
catatytic activity.
Regions
P4
and
P7
are both formed by
pairing
between conserved consensus seouences.
A sequence adjacent to P7 base
pairs
with the
sequence
that
contains the
reactive
G.
3',
f.
tla.
tt
I
V
---oH
G-P
€"
+
ilili;liii.
i'
,:
SeLf-spticing
occurs by transesterification
reactions
in which bonds
are exchanged direct:ty.
The bonds
that
have been
generated
at each stage
are
indi-
cated by the shaded
boxes.
All
group
I introrrs can
be organized
into a char-
acteristic second,rry
structure
with
nine helices
(Pl-P9).
i i,
',i"i ',.;
shows
a model
for the sec-
ondary structure
of the
Te tr ahymen
a inlr on.
\W o
of the base-pairerl
regions
are
generated
by
pair-
ing between con:;erved
sequence
elements
that
are common
to
g|oup I introns.
P4 is constructed
from the sequences
P and
Q;
P7
is formed from
sequences
R and
^i. The other
base-paired
regions
vary
in
sequenc(l
in individual
introns.
Muta-
tional analysis
ic entifies
an
intron
"core"
con-
taining P3, P4,
P6, and
P7, which
provides the
minimal region that
can undertake
a catalytic
reaction. The
leogths of
group I introns vary
widely, and the cr)nsensus
sequences
are
located
a considerable
distance
from
the actual
splice
junctions.
Some
of the
pairing reactions
are directly
involved in brinl;ing
the
splice
junctions
into a
conformation
that
supports
the
enzymatic
27.3 C;roup
I Introns
Form
a Characteristic
Secondary
Structure
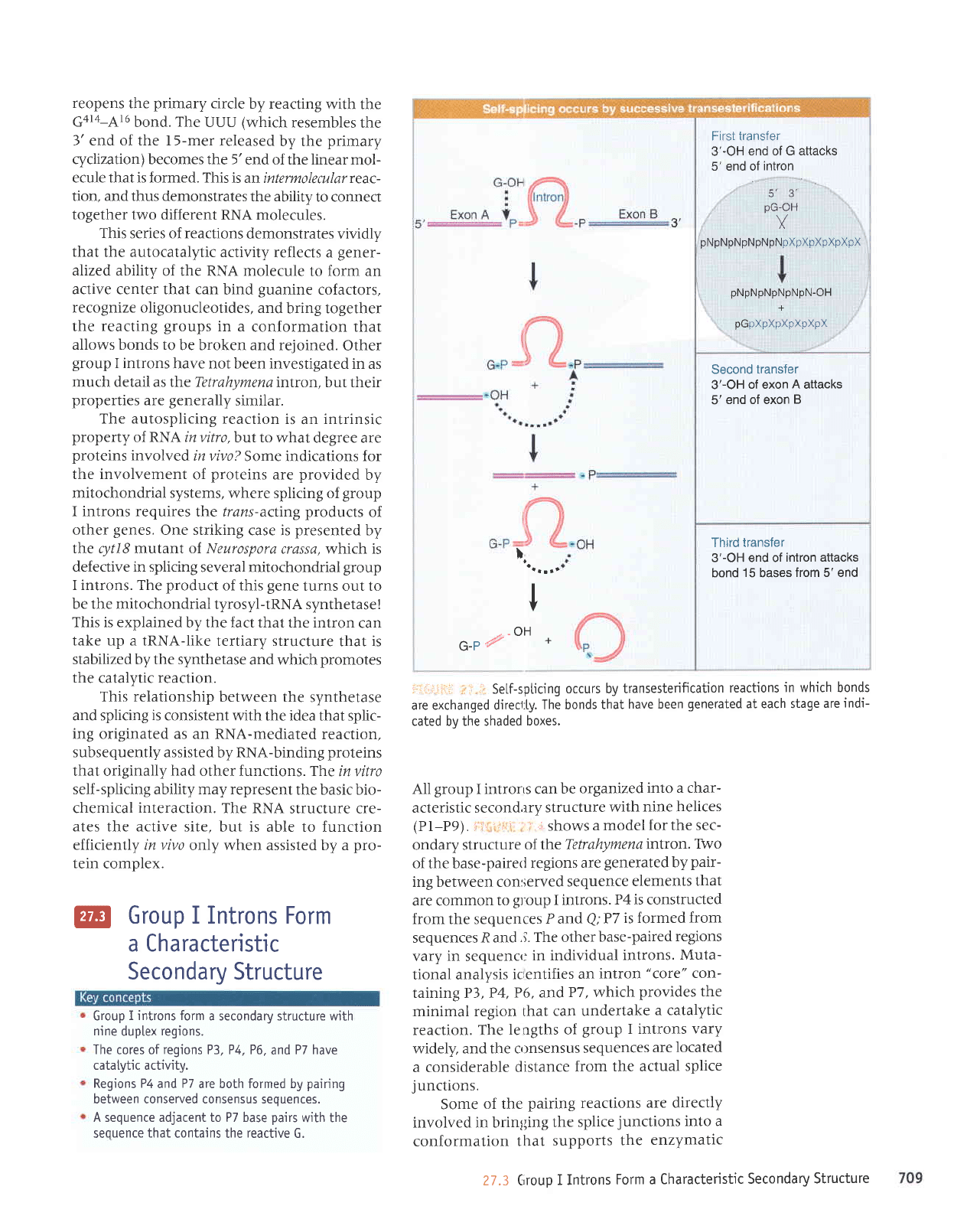
First transfer
3'-OH
end of G attacks
5' end of
intron
.
s',
3',
pG-OH
Y
pNpNpNpNpNpNpXpXFXpXpXpX
pNpNpNpNpNpN-OH
+
pGpXpXpXpXpXpX
Second transfer
3'-OH
of exon
A
attacks
5'
end of exon
B
Third transfer
3'-OH end of
intron attacks
bond
15
bases
from 5' end
3'-OH of
attacks
pArb
or
5'G..UUUpA1
6CCUpU2oUG
5'G..UUUpA
t
at+6gg
Cyclization
Reverse cvclization
-.^e I
H,O
ZrY"-
*
o'\
Linearization
products
414
1.19 RNA
lrans reaction
I
L-15 RNA
GalapAl6CCUpUzog6
uuupAl6ccupU20uG
P1
eron r
feoH
5, CUCUCU
First.
5,
3'GGGAGG
lransrer
3,
IGS
o
P8
J
5' UAGUC 3'
3'
AUCAG
5'
H
2 bp form
at 3' end
of intron
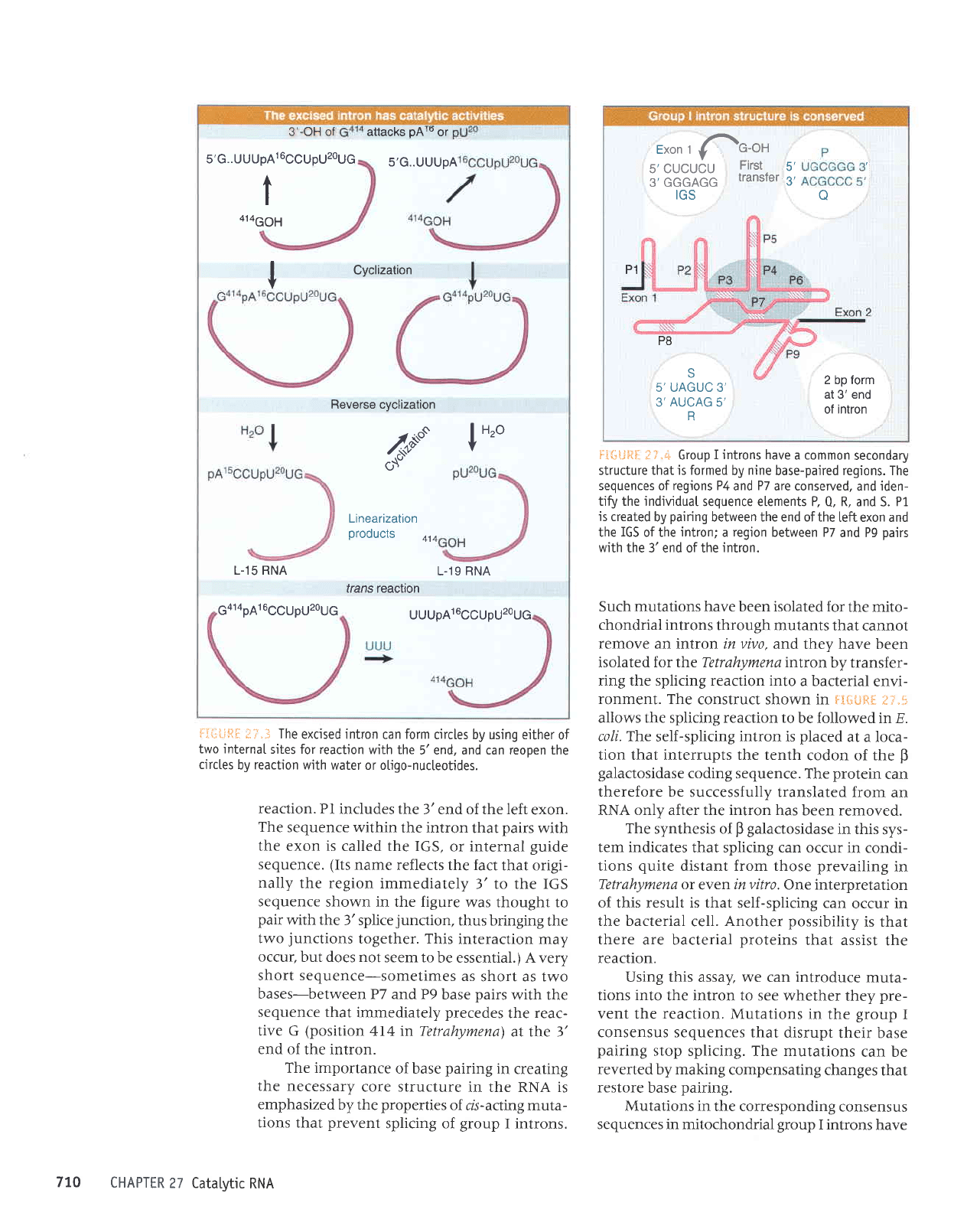
F:{iiigI
i]"3 The
excised intron
can
form
circles by using either
of
two internaI
sjtes for reaction
with the
5'end, and can reopen the
circtes
by reaction with
water or otigo-nucteotides.
reaction.
Pl includes the
3'end of the left
exon.
The
sequence within
the intron
that
pairs
with
the exon
is called the IGS,
or internal
guide
sequence. (Its
name
reflects the fact
that origi-
nally
the region
immediately
3' to the IGS
sequence
shown in the figure
was thought to
pair
with
the
3'splice
junction,
thus bringing
the
two
junctions
together. This
interaction may
occur,
but does not
seem to
be essential.) A very
short sequence-sometimes
as
short as two
bases-between
P7 and P9
base
pairs
with the
sequence that
immediately
precedes
the
reac-
tive G
(position
4I4 in Tetrahymena)
at the 3'
end of
the intron.
The importance
of base
pairing
in
creating
the
necessary
core structure
in the RNA is
emphasized
by the
properties
of
czi-acting muta-
tions that
prevent
splicing of
group
I introns.
CHAPTER
27 Catatytic
RNA
f,tililFqil
fF,4 Group
I introns have
a common
secondary
structure that is formed
by
nine
base-paired regions. The
sequences of regions P4
and
P7
are conserved,
and
iden-
tify the
jndividuat
sequence elements P,
Q,
R,
and S.
P1
js
created
by
pairing
between the end ofthe
left exon and
the
IGS
of the
intron;
a
region
between P7
and P9
pairs
with
the 3'end of the
intron.
Such mutations have been isolated for
the mito-
chondrial introns
through mutants
that cannot
remove an intron in vivo, and
they have been
isolated for the Tetrahymena intron
by transfer-
ring the splicing reaction into
a bacterial
envi-
ronment. The
construct shown in Fl{ilt*[
t3.*
allows the splicing reaction to
be followed in E.
coli. Tl;re
self
-splicing
intron
is
placed
at a loca-
tion that interrupts the tenth
codon of the
B
galactosidase
coding
sequence. The
protein
can
therefore be
successfully translated from
an
RNA only after the intron
has been removed.
The
synthesis of
B
galactosidase
in this
sys-
tem indicates
that splicing can
occur
in
condi-
tions
quite
distant from those
prevailing
in
Tetrahymena or
even
invitro.
One interpretation
of this result is that
self-splicing can
occur in
the bacterial
cell.
Another
possibility
is that
there
are bacterial
proteins
that assist
the
reaction.
Using this assay, we can introduce
muta-
tions into
the intron to see whether
they
pre-
vent
the
reaction.
Mutations
in the
group
I
consensus sequences that
disrupt
their base
pairing
stop splicing. The mutations
can be
reverted by making
compensating
changes
that
restore
base
pairing.
Mutations in
the corresponding
consensus
sequences in mitochondrial
group
I introns
have
770
