Schlick T. Molecular Modeling and Simulation: An Interdisciplinary Guide
Подождите немного. Документ загружается.


340 10. Nonbonded Computations
Linear Approximations to the PB Equation; Debye-H¨uckel Theory
For the case of dilute or moderate solutions (e.g., molar concentrations c
s
<
10
−3
M) and low fixed charge, it is possible to approximate the PB equation us-
ing results from Debye-H
¨
uckel theory (see [131, 275, 537, 547], for example, for
introductions). Specifically, a linearized approximation to the PB equation can be
used to represent the ionic atmosphere of a solute immersed in aqueous solution
and counterions.
A linearized version of the PB equation (eq. (10.58)) can be obtained by a
Taylor-series expansion of the Boltzmann factor with a truncation beyond the
first-order terms:
exp[−q
i
Φ(x)/k
B
T] ≈ 1 −[q
i
Φ(x)/k
B
T] . (10.59)
The linearized version is justified when
q
i
Φ(x) k
B
T ,
that is, when energies are much smaller than the thermal energy. This often
holds for monovalent electrolytes and weak source charges (dilute solution), as
mentioned above.
In the special case of spherical symmetry about the origin for the distribution
of charges, the potential Φ depends on the distance r of the point from the ori-
gin. Linearization of the PB equation yields the following linear second-order
differential equation for Φ(r) [537]:
6
∇
2
Φ(r)=κ
2
Φ(r) , (10.60)
where
κ
2
=
8πN
A
e
2
ρ
A
1000 k
B
T
c
s
. (10.61)
Here N
A
is Avogadro’s number, ρ
A
is the solvent density, e is the protonic charge
(4.803 ×10
−10
esu), and is the solvent dielectric constant. The ionic concentra-
tion c
s
is measured in molar units as a sum over all molar concentrations per liter
of solution, c
i
, associated with charges (or valences) q
i
: c
s
=
1
2
n
i
i=1
c
i
q
2
i
.
The linearized PB equation as expressed in eq. (10.60) with (10.61) can be
solved to determine the effective potential Φ at a distance r from a central ion.
Recall that the Coulomb potential q/(r) is produced at a point separated by dis-
tance r from an isolated central ion, which is represented as a uniform charged
sphere of radius a
r
in a medium of dielectric constant . The Debye-H¨uckel solu-
tion for the modified electrostatic potential representing the influence of the ionic
atmosphere is (e.g., [537]):
Φ(r)=B(κ)
q exp(−κr)
r
, (10.62)
6
We can also write
1
r
2
d
dr
r
2
d
dr
Φ(r)=κ
2
Φ(r).
10.6. Continuum Solvation 341
where B(κ) is the salt-dependent coefficient
B(κ)=exp(κa
r
)/(1 + κa
r
) . (10.63)
Thus, Debye-H¨uckel theory produces an effective electrostatic potential in
which the Coulomb interactions are screened by ions. The theory predicts the
range of electrostatic influence of a central ion to be the Debye screening length
κ
−1
.Inotherwords,κ
−1
is the characteristic distance of exponential screening.
From eq. (10.61), we see that the screening parameter κ is proportional to
the square root of the ionic concentration c
s
. For 1:1 electrolytes (monova-
lent:monovalent salts like NaCl),
κ ≈ 0.33
√
c
s
˚
A
−1
(10.64)
at room temperature (25
o
), with =78.5.
For dilute solutions, or κa
r
1,wehaveB(κ) ≈ 1; it follows that
the Coulomb screening — reduction of the unscreened potential by the factor
exp(−κr) — reflects the reduced effective charge ρ on the central ion due to
counterion accumulation. For physiological ionic strengths, such as 0.15M, the
Debye length is approximately 8
˚
A; this distance represents considerable damping
of Coulomb interactions.
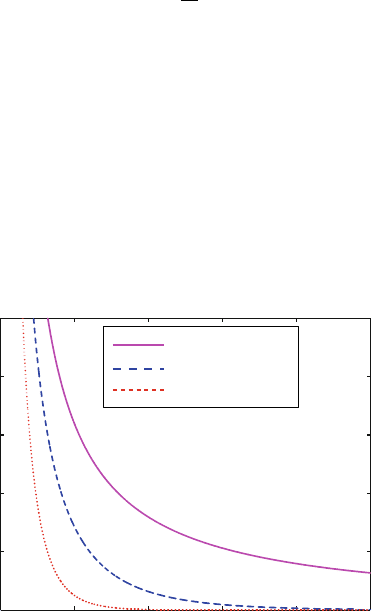
Figure 10.10 compares the Coulomb potential (1/r) to the screened Coulomb
potential (B(κ)exp[−κr]/r) for two values of κ corresponding to c
s
=0.15M
0 20 40 60 80 100
0
0.2
0.4
0.6
0.8
1
x 10
−3
Coulomb vs. Debye−Hückel Potentials
r (Ang)
Coulomb
DH,c
s
= 0.015 M
DH,c
s
= 0.15 M
Figure 10.10. Screened Coulomb potentials at two values of the Debye length. The
Coulomb potential (1/r) is compared to the screened Coulomb potential of form
B(κ)exp[−κr]/r as a function of distance r for two values of κ. These two values are
set according to eq. (10.64)forc
s
=0.15M and 0.015M monovalent salt concentrations,
with coefficients B(κ) computed according to eq. (10.63) (the coefficients are very close
to 1). The two κ values (0.128 and 0.040
˚
A, respectively) correspond to Debye lengths of
about 8 and 25
˚
A.
342 10. Nonbonded Computations
and 0.015M monovalent salt, where the Debye lengths are about 8 and 25
˚
A,
respectively.
The DH approximation has been applied to long DNA for exploring confor-
mational stability and mobility as a function of monovalent ionic concentrations
in the natural cellular environment (e.g., [1125, 1309]). Models are based on the
pioneering work of Stigter, who modeled DNA as charged cylinders [1223]and
reproduced the experimentally-observed dependence of the effective diameter of
DNA on salt concentration by a tail approximation to the PB equation. Extensions
of such DH approximations to macroscopic models of protein/DNA systems have
been described [109] and applied to model chromatin [108].
General Solutions to the Poisson-Boltzmann Equation
Many practical procedures have been developed to solve the PB equation nu-
merically, including solutions of its various approximations (e.g., [16, 80, 563,
610,813, 903, 1058, 1178]); see also references cited in [81], and [303,1023]for
applications to calculate charge distributions. The linearized approximations are
useful in many cases, as mentioned above. For high charge density (such as for
polyelectrolyte DNA) and high salt concentrations, the nonlinear PB version is
preferred.
Numerical solutions are typically obtained through finite-difference or finite-
element (or boundary element) methods. Both involve a discretization of the
(irregular) biomolecular domain in 3D, so that the potential, charge density, and
dielectric constant are defined at grid points ( is usually defined over broader
domain regions). The dielectric constant is assigned appropriate values depend-
ing on its proximity to the solute, though a two-dielectric model is typically used
— to distinguish the inside region, near the solute, from the outside region, far
from the solute. The finite-difference solution can be obtained iteratively by var-
ious linear algebra solvers (e.g., linear conjugate gradient method, Gauss-Seidel,
or Successive Over Relaxation methods) [16, 903,1058].
The resulting quality of the numerical solutions depends on the various assump-
tions made and settings used. The convergence of the solvers also depends on
such parameters as the grid size, initial charge and assignments, and algorithmic
parameters.
An analytical gradient minimization method based on a finite-element dis-
cretization for solving the PB equation has also been presented [429]. While the
DelPhi program uses a regular cubic grid, this gradient-based approach uses an
adaptive grid so as to include more grid points at the solvent-accessible surface,
where the dielectric value is changing rapidly. Overall, preliminary comparisons
show that the computational efficiency of both approaches is comparable [429];
the gradient-based method is also used for geometry optimization applications,
serving as an improvement to gas-phase molecular mechanics minimizations
since solvation effects are included.
Popular packages used in the biomolecular community are those developed at
research groups at Columbia University (DelPhi and GRASP) and the University
10.6. Continuum Solvation 343
of Houston (UHBD, now at UCSD, which includes segments for Brownian dy-
namics). The latter has been used to study huge macromolecular systems [81](see
also Figure 10.11).
The APBS (Adaptive PB Solver) program is also popular [81] and can be
used on its own or in conjunction with other molecular modeling packages like
CHARMM or AMBER, with results visualized with popular graphics programs
like VMD or PYMOL. In addition, web tools like PBEQ based on the PB solver
in CHARMM [610]orMark-US (a functional assessment tool for proteins) from
the Honig lab make applications easier to users by combining PB solvers with
interactive visualization modules.
Classical electrostatics solutions can provide useful information on charge
distributions around macromolecules (e.g., [303, 1023]), specific localization of
charged and polar groups in biomolecular systems (e.g., [790, 1253]), the shape
of molecular surfaces, and the relation between salt effects and conformational
changes (e.g., [1125]). Effects of the ionic atmosphere on intermolecular bind-
ing associations, such as between proteins and ligands, can also be analyzed
[565,1057].
Effective numerical packages such as DelPhi and UHBD, combined with ad-
vanced graphical rendering programs, offer valuable modeling tools for exploring
the effects of electrostatics and solvation on molecular structure and function.
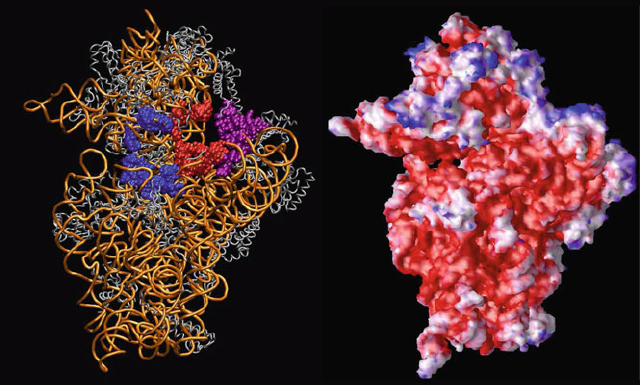
Figure 10.11. The electrostatic environment of the 30S ribosomal unit as computed by
the linear PB equation at 150 mM ionic strength with a solute dielectric of 2 and solvent
dielectric of 78.5 [81]. The protein (silver) and nucleic acid (gold) atoms are distinguished
on the left, with selected components of A, P, and E sites shown in blue, red, and purple
respectively. At right, the electrostatic potential mapped on the 30S surface is rendered in
blue (positive) and red (negative) to illustrate regions with values greater than 2.6 k
B
T/e
and less than −2.6 k
B
T/e, respectively.
344 10. Nonbonded Computations
The electrostatic analysis of a ribosomal unit is shown in Figure 10.11.(Seealso
analysis of the nucleosome core particle in Chapter 6).
Algorithmic Challenges
Recent algorithmic advances — by adaptive finite element methods [80,563]tai-
lored to multiprocessor systems [81] — allow solutions of the linear PBE for very
large systems of order one million atoms. Figure 10.11 illustrates the PB solution
for the 30S ribosome subunit.
A future goal in the field is repeated solution of the PBE, for example in the
course of a dynamic simulation.
11
Multivariate Minimization
in Computational Chemistry
Chapter 11 Notation
S
YMBOL DEFINITION
Matrices
A symmetric matrix, components {A
ij
}
B
k
approximation to Hessian inverse at x
k
(QN methods)
D
k
scaling matrix at step k of minimization method (trust
region approach)
H
Hessian matrix, components H
ij
(x) ≡ ∂
2
f(x)/∂x
i
∂x
j
I identity matrix
M
preconditioning matrix, related to H (TN methods)
M
k
preconditioning matrix at step k of TN method
U
k
QN low-rank update matrix at step k
Vectors
b, y constant vectors
e
j
unit vectors
g
gradient vector of f, components g
i
(x) ≡ ∂f(x)/∂x
i
g
k
gradient vector at x
k
(short hand for g(x
k
))
p
k
search vector at step k of minimization method
p
j
k
inner-loop CG iterate j for outer-loop search vector
p
k
(TN method)
r
residual vector defined in TN methods (Mz = r)
s
k
displacement vector at step k, x
k+1
− x
k
(QN method)
x
vector of n components {x
i
}
x
0
starting point vector for minimization
x
k
minimization iterate at step k of method
x
∗
local minimum point of objective function
y
k
gradient difference vector at QN step k, g
k+1
− g
k
z solution vector defined in TN methods (Mz = r)
T. Schlick, Molecular Modeling and Simulation: An Interdisciplinary Guide, 345
Interdisciplinary Applied Mathematics 21, DOI 10.1007/978-1-4419-6351-2
11,
c
Springer Science+Business Media, LLC 2010
346 11. Multivariate Minimization in Computational Chemistry
Chapter 11 Table (continued)
S
YMBOL DEFINITION
Scalars & Functions
a, b numbers
c
i
(x) constraint function i
c
r
small positive number (in TN methods)
f
0
constant (function value)
f(x)
objective function, dependent on vector x
h
small number (finite difference interval)
n
problem dimension
p
convergence order
q(x)
quadratic function
q
k
(s) quadratic model of objective function
r
k
residual norm at step k of TN methods
α
line search parameter for sufficient decrease condition
β
line search parameter for sufficient decrease of curvature
(also convergence ratio)
β
k
scheme-dependent scale parameter of search vector p
k
(CG and QN methods)
f
,
g
small positive numbers
m
small positive number, machine precision
η
k
forcing sequence in TN methods
λ
line search steplength
λ
t
trial line search steplength
ξ
variable in the neighborhood of x for a univariate
function f(x)
φ(λ)
polynomial of steplength λ
Δ
k
size bound in QN methods at step k
’Pon my word Watson, you are coming along wonderfully. We
have really done very well indeed. It is true that you have missed
everything of importance, but you have hit upon the method.
Arthur Conan Doyle (1859–1930), in A Case of Identity (1891).
Economic forecasting makes weather forecasting look like physics.
Ben Bernanke, 26 July 2009.
[Bernanke, the US Federal Reserve Chairman, spoke about the current
economic crisis in a town-hall forum in Kansas City, MO, hosted by the
PBS NewsHour’s Jim Lehrer.]
11.1. Ubiquitous Optimization: From Enzymes to Weather to Economics 347
11.1 Ubiquitous Optimization: From Enzymes
to Weather to Economics
Optimization is a fundamental component of molecular modeling. The deter-
mination of a low-energy conformation for a given force field can be the final
objective of the computation. It can also serve as a starting point for subsequent
calculations, such as molecular dynamics simulations or normal-mode analyses.
Both local and global optimization problems lie at the heart of numerous sci-
entific and engineering problems — from the biological and chemical disciplines
to architectural and industrial design to economics. Optimization is part of our
everyday life — responsible for our weather forecasts, flight planning, telephone
routing, microprocessor design, and the functioning of enzymes in our bodies.
11.1.1 Algorithmic Sophistication Demands Basic Understanding
The mathematical techniques developed to address these optimization problems
are just as robust and varied as the target problems themselves. The algorithmic
complexity of such techniques has led to many available computer programs that
require minimal input from the user (e.g., the starting point and a routine for
function evaluation).
However, the prudent user of these canned software modules — even within
standard molecular mechanics and dynamics packages — should understand the
fundamental structure of the optimization algorithms and associated performance
issues to make their application both efficient and correct, in terms of the physical
interpretations.
This chapter introduces key optimization concepts for this purpose. We also
highlight the fundamentals of local optimizers for large-scale nonlinear un-
constrained problems, an important optimization subfield relevant to biological
macromolecules. We describe the most promising approaches among them, and
discuss practical issues, such as parameter variations and termination criteria.
Of course, the latter are best learned by experimentation in the context of real
problems. To illustrate behavior for complex problems, some comparisons among
three competitive minimizers are also included, for molecular models minimized
in the molecular mechanics and dynamics program CHARMM.
11.1.2 Chapter Overview
Specifically, Section 11.2 introduces optimization fundamentals such as problem
formulation and terminology. Section 11.3 describes the basic algorithmic frame-
work of iterative minimization protocols (based on line search and trust region
methods); it also discusses convergence criteria and line search procedures and
introduces the key concept of descent directions.
In Section 11.4, we present the Newton method, including a historical per-
spective, and one-dimensional implementations for nonlinear equations as well
348 11. Multivariate Minimization in Computational Chemistry
as optimization. This presentation familiarizes readers with the Newton method
framework — the basis for formulating many other optimization methods — and
with performance and convergenceissues relevant to minimization of multivariate
functions.
In Section 11.5, we mention effective methods for large-scale nonlinear op-
timization, namely quasi-Newton (QN), nonlinear conjugate gradient (CG), and
truncated Newton (TN) schemes. Section 11.6 outlines available software and
presents comparative performance in CHARMM for two molecular models
(a small model system and a protein). Finally, in Sections 11.7 and 11.8,we
summarize recommendations to optimization practitioners and offer a future
perspective to field developments.
For details, as well as for other categories of the rich and exciting field of
optimization, I refer readers to classic texts [297, 407, 459, 789, 918], some re-
views [417, 915, 1104, 1108, 1131], and a perspective [1390]. In recent years,
optimization methods have become increasingly important in material science and
nanotechnology, for finding the favored structure of materials, including biologi-
cal systems. Such problems are typically solved by global optimization problems,
as recently reviewed [196].
11.2 Optimization Fundamentals
The methods for solving an optimization task depend on the problem classifica-
tion. Since the value of the independent variable that maximizes a function f also
minimizes the function −f, it suffices to deal with minimization.
The optimization problem is classified according to the type of indepen-
dent variables involved (real, integer, mixed), the number of variables (one,
few, many), the functional characteristics (linear, least squares, nonlinear, non-
differentiable, separable, etc.), and the problem statement (unconstrained, subject
to equality constraints, subject to simple bounds, linearly constrained, nonlin-
early constrained, etc.). For each category, suitable algorithms exist that exploit
the problem’s structure and formulation.
11.2.1 Problem Formulation
For a vector x of n components {x
i
}, we write the minimization problem as:
min
x
{f(x)} , x ∈D, (11.1)
where f is the objective function and D is a given region (which can be the entire
Euclidean space
n
). The problem can be subject to m constraints, which can be
written more generally as a combination of equality and inequality constraints:
c
i
(x)=0 fori =1,...,m
,
c
i
(x) ≤ 0fori = m
+1,...,m. (11.2)

11.2. Optimization Fundamentals 349
This general formulation can be obtained for problems with bound constraints in
the form
c
i
(x)=x
i
,
where x
i
is the ith component of the vector x, or for problems with two-sided
constraints such as
l
i
≤ c
i
(x) ≤ u
i
.
In this chapter, we only cover unconstrained optimization formulations. For a
comprehensive review of interior methods for continuous nonlinear optimization
problems subject to constraints, see [417].
11.2.2 Independent Variables
In most computational chemistry problems, x is a real vector in Euclidean space,
i.e., x ∈
n
,andf defines a transformation to a real number,i.e., f(x):
n
→.
When the components of x are integers, the optimization problem is classified
as integer programming.Whenx is a mixture of real and integer variables, the
problem is of mixed-integer programming type. Common examples of integer pro-
gramming are network optimization and the ‘traveling salesman problem’,
1
also
classified as combinatorial optimization. See [1390], for example, and references
cited therein.
11.2.3 Function Characteristics
The nature of the function f is the next step in problem classification. Many appli-
cation areas such as finance and management-planning tackle linear or quadratic
objective functions.
Linear and Quadratic Functions
Linear objectives can be written in vector form as
f(x)=b
T
x + f
0
, (11.3)
where b is a column vector of dimension n,andf
0
is a scalar. Quadratic objective
functions can be expressed as
f(x)=x
T
Ax + b
T
x + f
0
, (11.4)
1
The notorious ‘traveling salesman’ problem seeks to find the optimal travel route that covers
a given number of cities, each one only once, and returning to the home town. Visually, imagine
drawing such a route on a map, where each city k for k =0,...,n is designated by coordinates
{x
k
,y
k
}. The connected route started at {x
0
,y
0
} covers each city and returns to the original point.
Though simple to envision, there are clearly many such routes, and the number of combinations that
connect all these cities grows steeply with n. This problem in fact belongs to a class of very difficult
problems (known as NP-complete) for which no polynomial-complexity algorithm is known (i.e., the
computational time for an exact solution of this problem increases exponentially with n).
