Vidakovic B. Statistics for Bioengineering Sciences: With Matlab and WinBugs Support
Подождите немного. Документ загружается.

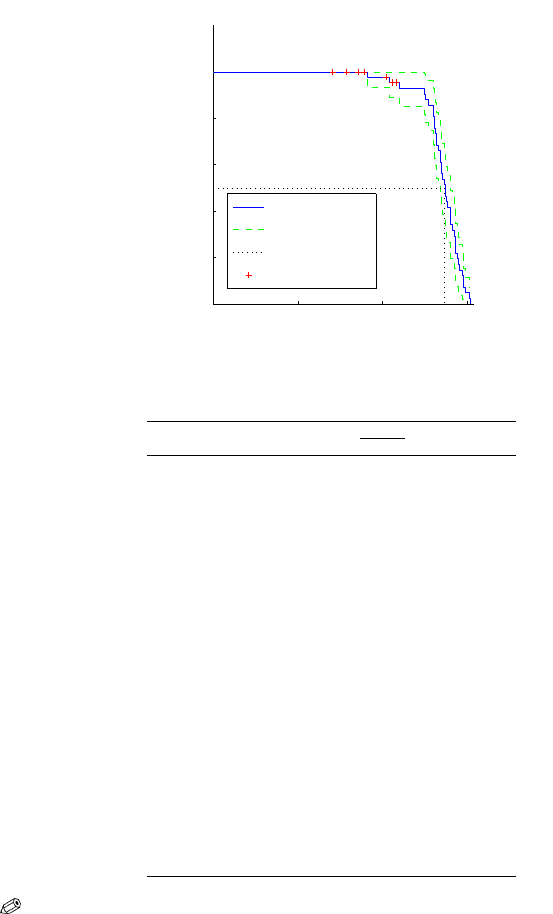
18.3 Inference with Censored Observations 711
censor=[ones(1,41),zeros(1,7)];
[kmest,sortdat,sortcen]= KMcdfSM(data’,censor’,0);
plot(sortdat,1-kmest,’k’);
The table below shows how the Kaplan–Meier estimator is calculated for the
first 16 measurements, which includes 7 censored observations. Figure 18.3
shows the estimated survival function for the cord strength data.
0 20 40 60
0
0.2
0.4
0.6
0.8
1
ˆ
S(t)
Time
Data
95% CI
Med T 54.656
Censored
Fig. 18.3 Kaplan–Meier estimator cord strength (in coded units).
Uncensored x
j
m
j
d
j
m
j
−d
j
m
j
1 −F
K M
(x
j
)
26.8 48 0 1.000 1.000
29.6 47 0 1.000 1.000
33.4 46 0 1.000 1.000
35.0 45 0 1.000 1.000
1 36.3 44 1 0.977 0.977
40.0 43 0 1.000 0.977
2 41.7 42 1 0.976 0.954
41.9 41 0 1.000 0.954
42.5 40 0 1.000 0.954
3 43.9 39 1 0.974 0.930
4 49.9 38 1 0.974 0.905
5 50.1 37 1 0.973 0.881
6 50.8 36 1 0.972 0.856
7 51.9 35 1 0.971 0.832
8 52.1 34 1 0.971 0.807
9 52.3 33 2 0.939 0.758
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.
.

712 18 Inference for Censored Data and Survival Analysis
18.3.3 Comparing Survival Curves
In clinical trials it is often important to compare survival curves calculated
for cohorts undergoing different treatments. Most often one is interested in
comparing the new treatment to the existing one or to a placebo. In comparing
two survival curves we are testing whether the corresponding hazard func-
tions h
1
(t) and h
2
(t) coincide:
H
0
: h
1
(t) = h
2
(t) vs H
1
: h
1
(t) >, 6=,< h
2
(t).
The simplest comparison involves exponential lifetime distributions where
the comparison between survival/hazard functions is simply a comparison of
constant rate parameters. The statistic is calculated using logarithms of haz-
ard rates as
Z
=
logλ
1
−log λ
2
p
1/k
1
+1/k
2
,
where k
1
and k
2
are numbers of observed (uncensored) survival times in the
two comparison groups. Now, the inference relies on the fact that statistic Z is
approximately standard normal.
Example 18.8. In Example 18.5 the rate for immunoperoxidase-positive pa-
tients was larger than that of immunoperoxidase-negative patients? Was this
difference significant?
z = (log(lambdahat1)-log(lambdahat2))/sqrt(1/k1+1/k2) %-2.5763
p = normcdf(z) %0.0050
As is evident from the code, the hazard rate λ
2
(and, in the case of exponential
distribution, hazard function) for the immunoperoxidase-positive patients is
significantly larger than the rate for negative patients,
λ
1
, with a p-value of
1/2 percent.
LogRank Test. Tests for comparing survival functions in nonparamet-
ric fashion use Hanszel–Mantel theory on survival data represented as 2
×2
tables. Another popular term is logrank test.
Let (r
11
, d
11
), (r
12
, d
12
),... , (r
1k
, d
1k
) be the number of people at risk and
the number of people who died at times t
11
, t
12
,... , t
1k
in the first cohort,
and (r
21
, d
21
), (r
22
, d
22
),... , (r
2m
, d
2m
) be the number of people at risk and the
number of people who died at times t
21
, t
22
,... , t
1m
in the second cohort. We
merge the two data sets together with the corresponding times. Thus there will
be D
= k +m time points if there are no ties, and each time point corresponds
to a death from either the first or second cohort. For example, if times of events
in the first sample are 1, 4, and 10 and in the second 2, 3, 7, and 8, then in the
merged data sets the times will be 1, 2, 3, 4, 7, 8, and 10.

18.3 Inference with Censored Observations 713
For a time t
i
from the merged data set, let r
1i
and r
2i
correspond to the
number of subjects at risk in cohorts 1 and 2, respectively, and let r
i
= r
1i
+r
2i
be the number of subjects at risk in the combined sample. Analogously, let d
1i
,
d
2i
, and d
i
= d
1i
+d
2i
be the number of events at time t
i
.
Then, if H
0
: h
1
(t) = h
2
(t) is true, then d
1i
has a hypergeometric distribu-
tion with parameters (r
i
, d
i
, r
1i
).
Event No event At risk
Treatment 1 d
1i
r
1i
−d
1i
r
1i
Treatment 2 d
2i
r
2i
−d
2i
r
2i
Merged d
i
r
i
−d
i
r
i
Since d
1i
∼H G (r
i
, d
i
, r
1i
), the expectation and variance of d
1i
are
Ed
1i
= r
1i
×
d
i
r
i
,
Var (d
1i
) =
r
i1
r
i
µ
1 −
r
i1
r
i
¶µ
r
i
−d
i
r
i
−1
¶
d
i
.
Note that in the terminology of the Kaplan–Meier estimator, the number
of subjects with no event at time t
i
is equal to r
i
−d
i
= r
i+1
+`
i
, where `
i
is
the number of subjects censored in the time interval (t
i−1
, t
i
) and r
i+1
is the
number of subjects at risk at the beginning of the subsequent interval (t
i
, t
i+1
).
The test statistic for testing H
0
: h
1
(t) = h
2
(t) against the two-sided alter-
native H
1
: h
1
(t) 6= h
2
(t) is
χ
2
=
¡
P
D
i
=1
(
d
i1
−E(d
1i
)
)
¢
2
P
D
i
=1
Var (d
1i
)
,
which has a
χ
2
-distribution with 1 degree of freedom. The continuity cor-
rection 0.5 can be added to the numerator of the
χ
2
-statistic as (|
P
D
i
=1
(d
i1
−
E
(d
1i
))|−0.5)
2
when the sample size is small. If the statistic is calculated as
chi2, then its large values are critical and the p-value of the test is equal to
1-chi2cdf(chi2,1).
If the alternative is one-sided, H
1
: h
1
(t) < h
2
(t) or H
1
: h
1
(t) > h
2
(t), then
the preferable statistic is
Z
=
P
D
i
=1
(
d
i1
−E(d
1i
)
)
q
P
D
i
=1
Var (d
1i
)
714 18 Inference for Censored Data and Survival Analysis
and the p-values are normcdf(Z) and 1-normcdf(Z), respectively. A more gen-
eral statistic is of the form
Z
=
P
D
i
=1
W(t
i
)
(
d
i1
−E(d
1i
)
)
q
P
D
i
=1
W
2
(t
i
)Var (d
1i
)
,
where W(t
i
) = 1 (as above), W(t
i
) = r
i
(Gehan’s statistic), and W(t
i
) =
p
r
i
(Tarone–Ware statistic).
Example 18.9. Histiocytic Lymphoma. The data (from McKelvey et al.,
1976; Armitage and Berry, 1994) given below are survival times (in days) since
entry to a trial by patients with diffuse histiocytic lymphoma. Two cohorts of
patients are considered: (1) with stage III and (2) with stage IV of the disease.
The observations with + are censored.
Stage 6 19 32 42 42 43+ 94 126+ 169+ 207 211+
III
227+ 253 255+ 270+ 310+ 316+ 335+ 346+
Stage 4 6 10 11 11 11 13 17 20 20 21
IV
22 24 24 29 30 30 31 33 34 35 39
40 41+ 43+ 45 46 50 56 61+ 61+ 63 68
82 85 88 89 90 93 104 110 134 137 160+
169 171 173 175 184 201 222 235+ 247+ 260+ 284+
290+ 291+ 302+ 304+ 341+ 345+
Using a logrank test we will assess the equality of the two survival curves.
Figure 18.4 and table
%----------------------------------------------------------
% L S.E. z p-value alpha
%----------------------------------------------------------
% 8.65786 3.35325 2.43283 0.01498 0.050
%----------------------------------------------------------
% The survival functions are statistically different
are outputs from logrank.m (Cardillo, 2008).
18.4 The Cox Proportional Hazards Model
We often need to take into account that survival is influenced by one or more
covariates, which may be categorical (such as the kind of treatment a patient
received) or continuous (such as the patient’s age, weight, or the dosage of
a drug). For simple situations involving a single factor with just two values
(such as drug versus placebo) we discussed a method for comparing the sur-
vival curves for the two groups of subjects. But for more complicated situations
a regression-type model that incorporates the effect of each predictor on the
shape of the survival curve is needed.
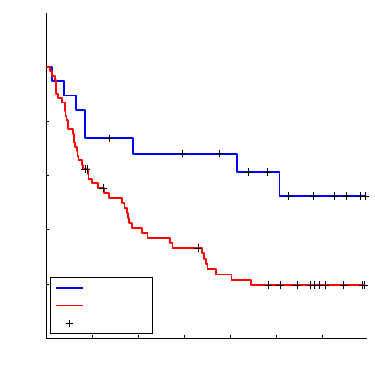
18.4 The Cox Proportional Hazards Model 715
0 50 100 150 200 250 300
0
0.2
0.4
0.6
0.8
1
Estimated Survival Functions
Time
Group 1
Group 2
Censored
Fig. 18.4 Kaplan–Meier estimators of the two survival functions. Here group 1 corresponds
to Stage III and group 2 to Stage IV cohorts.
Assume that the log hazard for subject i can be modeled via a linear rela-
tionship:
log h(t, x
i
) =β
0
+β
1
x
1,i
+···+β
p
x
p,i
,
where x
i
= x
1,i
,... , x
p,i
is p-dimensional vector of covariates associated with
subject i. The Cox model assumes that
β
0
is a log baseline hazard, log h
0
(t) =
β
0
, i.e., log hazard for a “person” for whom all covariates are 0 (Cox, 1972; Cox
and Oakes, 1984). Alternatively, one can set the baseline hazard to correspond
to a person for whom all covariates are averages of covariates from all subjects
in the study. For the Cox model,
log h(t, x
i
) =log h
0
(t) +β
1
x
1,i
+···+β
p
x
p,i
,
or, equivalently,
h(t, x
i
) = h
0
(t) ×exp{β
1
x
1,i
+···+β
p
x
p,i
} = h
0
(t) ×exp{x
0
i
β}.
Inclusion of an intercept would lead to nonidentifiability because
h
0
(t) ×exp{x
0
i
β} =(h
0
(t)e
−α
) ×exp{α +x
0
i
β}.
This allows for some freedom in choosing the baseline hazard.
For two subjects, i and j, the ratio of hazard functions
716 18 Inference for Censored Data and Survival Analysis
h(t, x
i
)/h(t, x
j
) =exp{(x
0
i
−x
0
j
)β}
is free of t, motivating the name proportional. Also, for a subject i,
S(t, x
i
) =
(
S
0
(t)
)
exp{x
0
i
β}
,
where S
0
(t) is the survival function corresponding to the baseline hazard h
0
(t).
This follows directly from (18.1) and H(t, x
i
) = H
0
(t)exp{x
0
i
β}.
In MATLAB,
coxphfit fits the Cox proportional hazards regression model,
which relates survival times to predictor variables. The following example
uses
coxphfit to fit Cox’s proportional hazards model.
Example 18.10. Mayo Clinic Trial in PBC. Primary biliary cirrhosis (PBC)
is a rare but fatal chronic liver disease of unknown cause, with a prevalence of
about 1/20,000. The primary pathologic event appears to be the destruction of
interlobular bile ducts, which may be mediated by immunologic mechanisms.
The PBC data set available at StatLib is an excerpt from the Mayo Clinic
trial in PBC of the liver conducted between 1974 and 1984. From a total of
424 patients that met eligibility criteria, 312 PBC patients participated in the
double-blind, randomized, placebo-controlled trial of the drug D-penicillamine.
Details of the trial can be found in Markus et al. (1989).
Survival statuses were recorded for as many patients as possible until July
1986. By that date, 125 of the 312 patients had died and 187 were censored.
The variables contained in the data set
pbc.xls|dat are described in the
following table:
casen = pbc(:,1); %case number 1-312
lived = pbc(:,2); %days lived (from registration to study date)
indicatord = pbc(:,3); %0 censored, 1 death
treatment = pbc(:,4); %1 - D-Penicillamine, 2 - Placebo
age = pbc(:,5); %age in years
gender = pbc(:,6); %0 male, 1 female
ascites= pbc(:,7); %0 no, 1 yes
hepatomegaly=pbc(:,8); %0 no, 1 yes
spiders = pbc(:,9); %0 no, 1 yes
edema = pbc(:,10); %0 no, 0.5 yes/no therapy, 1 yes/therapy
bilirubin = pbc(:,11); %bilirubin [mg/dl]
cholesterol = pbc(:,12); %cholesterol [mg/dl]
albumin = pbc(:,13); %albumin [gm/dl]
ucopper =pbc(:,14); %urine copper [mg/day]
aphosp =pbc(:,15); %alcaline phosphatase [U/liter]
sgot = pbc(:,16); %SGOT [U/ml]
trig =pbc(:,17); %triglycerides [mg/dl]
platelet = pbc(:,18); %# platelet count [#/mm^3]/1000
prothro = pbc(:,19); %prothrombin time [sec]
histage = pbc(:,20); %hystologic stage [1,2,3,4]
To illustrate the CPH model, in this example we selected four predictors
and formed a design matrix X as

18.4 The Cox Proportional Hazards Model 717
X = [treatment age gender edema];
The treatment has two values, 1 for treatment by D-penicillamine and 2
for placebo. The variable
edema takes three values: 0 if no edema is present,
0.5 when edema is present but no diuretic therapy was given or edema re-
solved with diuretic therapy, and 1 if edema is present despite administration
of diuretic therapy.
The variable
lived is the lifetime observed or censored, a censoring vector
is
1-indicatord, and a baseline hazard is taken to be a hazard for which all
covariates are set to 0.
[b,logL,H,stats] = coxphfit(X,lived,...
’censoring’,1-indicatord,’baseline’,0);
The output H is a two-column matrix as a discretized cumulative hazard
estimate. The first column of
H contains values from the vector lived, while the
second column contains the estimated baseline cumulative hazard evaluated
at
lived.
To illustrate the model, we selected two subjects from the study to find
survival curves corresponding to their covariates. Subject #100 is a 51-year-old
male with no edema who received placebo while subject #275 is a 38-year-old
female with no edema who received D-penicillamine treatment.
X(100,:) %2.0000 51.4689 0 0
% placebo; 51 y.o.; male; no edema;
X(275,:) %1.0000 38.3162 1.0000 0
% D-Penicillamine; 38 y.o.; female; no edema;
First we find cumulative hazards at the mean values of predictors, as well
as for subjects #100 and #275, as
H(t,
x) = H
0
(t) ×exp{β
1
x
1
+···+β
4
x
4
},
H(t, x
i
) = H
0
(t) ×exp{β
1
x
1,i
+···+β
4
x
4,i
}, i =100, 275
Hmean(:,2) = H(:,2) .
*
exp(mean(X)
*
b); %c.haz. average
Hsubj100(:,2) = H(:,2) .
*
exp(X(100,:)
*
b); %subject #100
Hsubj275(:,2) = H(:,2) .
*
exp(X(275,:)
*
b); %subject #275
Here, the estimators of coefficients β
1
,... , β
4
are
b’
% 0.0831 0.0324 -0.3940 2.2424
Note that the treatment coefficient 0.0831 >0 indicates that, given all other
covariates fixed, the placebo increases the risk over the treatment. Also note
that
age and edema statuses also increase the risk, while the risk for female
subjects is smaller.
Next, from cumulative hazards we find survival functions
Smean = exp(-Hmean(:,2));
Ssubj100 = exp(-Hsubj100(:,2));
Ssubj275 = exp(-Hsubj275(:,2));
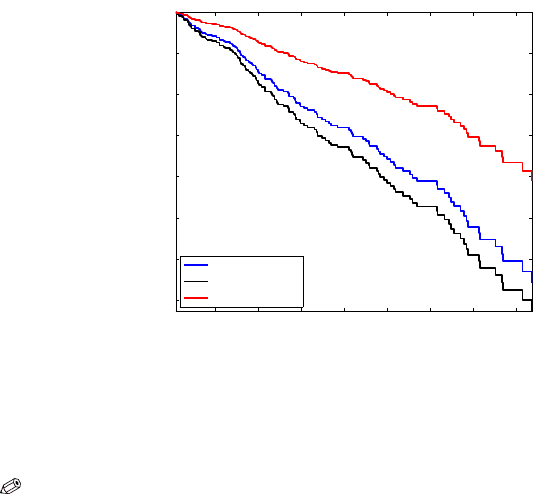
718 18 Inference for Censored Data and Survival Analysis
The subsequent commands plot the survival curves for an “average” subject
(blue), as well as for subjects #100 (black) and #275 (red), see Fig. 18.5.
stairs(H(:,1),Smean,’b-’,’linewidth’,2)
hold on
stairs(H(:,1),Ssubj100,’k-’)
stairs(H(:,1),Ssubj275,’r-’)
xlabel(’$t$ (days)’,’Interpreter’,’LaTeX’)
ylabel(’$
\hat S(t)$’,’Interpreter’,’LaTeX’)
legend(’average subject’,’subject #100’, ’subject #275’, 3)
axis tight
500 1000 1500 2000 2500 3000 3500 4000
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1
t (days)
ˆ
S(t)
average subject
subject #100
subject #275
Fig. 18.5 Cox model for survival curves for a “subject” with average covariates, subject #100
(51-year-old male on placebo, no edema), and subject #275 (38-year-old female on treatment,
no edema).
18.5 Bayesian Approach
We will focus on parametric models in which the lifetime distributions are
specified up to unknown parameters. The unknown parameters will be as-
signed prior distributions and the inference will proceed in a Bayesian fash-
ion. Nonparametric Bayesian modeling of survival data is possible; however,
the methodology is advanced and beyond the scope of this text. For a compre-
hensive coverage see Ibrahim et al. (2001).
Let survival time T have distribution f (t
|θ), where θ is unknown parame-
ters. For t
1
,... , t
k
observed and t
k+1
,... , t
n
censored times, the likelihood is

18.5 Bayesian Approach 719
L(θ|t
1
,... , t
n
) =
k
Y
i=1
f (t
i
|θ)×
n
Y
i=k+1
S(t
i
|θ).
If the prior on
θ is π(θ), then the posterior is
π(θ|t
1
,... , t
n
) ∝ L(θ|t
1
,... , t
n
) ×π(θ).
The Bayesian estimator of hazard is
ˆ
h
B
(t) =
Z
h(t|θ)π(θ|t
1
,... , t
n
)dθ
and the survival function is
ˆ
S
B
(t) =
Z
S(t|θ)π(θ|t
1
,... , t
n
)dθ.
Example 18.11. In Example 18.4 we showed that for the exponential lifetime
in the presence of censoring, the MLE for
λ is
ˆ
λ = k/
n
X
i=1
t
i
,
where k is the number of uncensored data and
P
n
i
=1
t
i
is the sum of all ob-
served and censored times and that the likelihood was L(
λ) =λ
k
exp{−λ
P
n
i
=1
t
i
}.
If a gamma
G a(α, β) prior on λ is adopted, π(λ) ∝λ
α−1
exp{−βλ}, then the con-
jugacy leads to the posterior
π(λ|t
1
,... , t
n
) ∝λ
k+α−1
exp{−(β +
n
X
i=1
t
i
) λ},
from which the Bayes estimator of
λ is the posterior mean,
ˆ
λ
B
=
k +α
β +
P
n
i
=1
t
i
.
One can show that the posterior predictive distribution of future failure time
t
n+1
is
f (t
n+1
|t
1
,... , t
n
) =
Z
∞
0
λe
−λt
n+1
×π(λ|t
1
,... , t
n
)dλ
=
(k +α)
¡
β +
P
n
i
=1
t
i
¢
α+k
¡
β +
P
n
i
=1
t
i
+t
n+1
¢
α+k+1
, y
n+1
>0.
This distribution is known as an inverse beta distribution.
The Bayes estimator of hazard function coincides with the Bayes estimator
of
λ,

720 18 Inference for Censored Data and Survival Analysis
ˆ
h
B
(t) =
k +α
β +
P
n
i
=1
t
i
,
while the Bayes estimator of the survival function is
ˆ
S
B
(t) =
Ã
1 +
t
β +
P
n
i
=1
t
i
!
−(k+α)
.
The expression for
ˆ
S
B
(t) can be derived from the moment-generating function
of a gamma distribution.
When the posterior distribution is intractable, one can use WinBUGS.
18.5.1 Survival Analysis in WinBUGS
WinBUGS uses two arrays to define censored observations: observed (uncen-
sored) times and censored times. For example, an input such as
list(times = c(0.5, NA, 1, 2, 6, NA, NA),
t.censored = c(0, 0.9, 0, 0, 0, 9, 12))
corresponds to times {0.5,0.9+,1,2,6, 9+, 12+}.
In WinBUGS, direct time-to-event modeling is possible with exponential,
Weibull, gamma, and log-normal densities.
There is a multiplier I that is used to implement censoring. For exam-
ple, if Weibull
dweib(r, mu) observations are on the input, the multiplier
exceeded time
t.censored[i]:
t[i] ~ dweib(r, mu) I(t.censored[i],)
Example 18.12. Bayesian Immunoperoxidase and BC. In Example 18.5
MLEs and confidence intervals on
λ
1
and λ
2
were found. In this example we
find Bayes estimators and credible sets.
Before discussing the results, note that when observations are censored,
their values are unknown parameters in the Bayesian model and predictions
can be found. On the other hand, all censored observations need to be ini-
tialized, and the initial values should exceed the censoring times. Here is the
WinBUGS program that estimates
λ
1
and λ
2
.
model{
for(i in 1:n1) {
ImmPeroxNeg[i] ~ dexp(lam1) I(CensorIPN[i], )
For uncensored data, t.censored[i] = 0. The above describes right-censoring.
Left-censored observations are modeled using the multiplier
I(,t.censored[i]).
