Feny? D. (Ed.) Computational Biology
Подождите немного. Документ загружается.

120 Gronwald and Kalbitzer
resolution (number of data points). This is the case with the
cosine filter as well as the Lorentzian-to-Gaussian filter (here
as an artifact resulting from the definitions provided by
TOPSPIN) (also see Subheading 3.3).
7. Application of an unshifted sine filter removes the volume
information (the volume of all cross peaks is zero) (also see
Subheading 3.3).
8. Do a careful baseline correction of all spectra before starting
with the data evaluation (also see Subheading 3.3.1).
9. Apply the different processing methods that your software
provides sequentially, e. g., a back prediction of the first data
points in the time domain, followed by appropriate filtering
and Fourier transform of the time domain data and a poly-
nomial baseline correction in the frequency domain (also see
Subheadings 3.3 and 3.3.1).
10. Be careful that some of the different processing func tions do
not work properly with oversampled data that have a different
time domain data structure (also see Subheadings 3.3
and 3.3.1).
11. Most carefully prepare your peak list since the quality
of the peak list is the most important single factor influen-
cing the outcome of any automated procedure (also see
Subheadings 3.3.2, 3.4, and 3.4.1).
12. Do not remove significant weak peaks by a too high peak
picking threshold (in AUREMOL the threshold is set auto-
matically) (also see Subheadings 3.4 and 3.4.1).
13. Bayesian methods are rather powerful to discriminate between
true signals and noise and artifacts, but they can only decide
on the basis of the information available. Important
information is the number of expected cross peaks that
depends on the pulse sequence and the sample composition.
Choose the final probability cutoff such that the number of
experimental cross-peaks corresponds to the expectation.
A factor of two is still acceptable by many routines (also see
Subheadings 3.4 and 3.4.1).
14. Try to understand the principles of the integration software
used (also see Subheading 3.4.2).
15. The iterative segmentation software that facilitates the
signal integration of AUREMOL requires the definition of a
maximum integration area. This has to be defined large
enough, otherwise the integrals will be erroneous (also see
Subheading 3.4.2).
16. For an efficient computing such a parameter is also (inherently)
present in many other routines (read the manual!) (also see
Subheading 3.4.2).
121
Automated Protein NMR Structure Determination in Solution
17. For all sequential assignment methods, the obtained result
strongly depends on the quality of the peak lists (see above),
since artifact peaks and missing peaks lead to wrong or
ambiguous connectivities. In most cases, connectivity infor-
mation solely based on Ca’s is not sufficient for obtaining
unambiguous assignments. Therefore, it is highly recom-
mended to additionally provide information for the Cb ’s as
well. In case that a nonexhaustive routine like simulated
annealing is used, it is generally recommended to perform the
routines several times and to analyze the results for discrepan-
cies between the different runs (also see Subheading 3.5).
18. The method that has been implemented in AUREMOL for
side-chain assignment works directly on the NOESY spectra;
therefore, the preparation of peak lists is not critical (also see
Subheading 3.6).
19. Since for the side-chain assignments the backbone assign-
ments are a strong source of information (when available), be
careful that the chemical shifts given fit optimally to the
NOESY spectra used. A typical effect that should be corrected
for is the shift introduced by using TROSY methods (also see
Subheading 3.6).
20. For automated NOE assignment, carefully prepare the
peak list. Be sure that the peak picking threshold is low
enough to retain the weak long range peaks. Set the proba-
bility cutoff at a value that the number of peaks is smaller
than twice the number of expected cross peaks (also see
Subheading 3.7).
21. Before starting the automated NOE assignment, be sure that
the chemical shift table corresponds closely to the conditions
used for the recording of the NOESY-spectrum. Apply the
chemical shift optimization routine AUREMOL-SHIFTOPT
(78) that adapts the shift table to the actual spectrum (also
see Subheading 3.7).
22. Adapt the other parameters (especially the search distance
range) to the iteration cycle (see also Subheadings
3.7.1–3.7.4).
23. For many of the parameters, default values are recommended
by the program. For an additional detailed description of the
various parameters, please refer to the AUREMOL manual
(see also Subheadings 3.7.1–3.7.4).
24. Note that upper and lower bounds are added to and subtracted
from the distance dist that was obtained from the individual
cross-peak volume. Relatively large upper bounds were selected
for the first cycles of structure calculations to ensure that
the influence of erroneous assignments is limited. Upper
bound definition was taken from Linge et al., 2001 (79).

122 Gronwald and Kalbitzer
Lower bounds were selected in a way that the minimal distance
between two protons corresponds to the sum of their van der
Waals radii (see also Subheadings 3.7.1–3.7.4).
25. Please note that for the last iteration (iteration 6) the option
“assign all peaks” has been switched on, which leads to an
increased number of NOE assignments (should only be done
in the last iteration), since still ambiguous NOEs are assigned
to the solution that correspond to the smallest distance in the
trial structure, since this contribution will explain most of the
crosspeak volume. It is clear that for truly overlapping signals
the minor signal component will be neglected, which, in turn,
will lead to an underestimation of the resulting restraint
distance of the major component. However, in most cases,
this deviation is rather small and within the measurement
error. For the relaxation matrix-based restraint generation
(see below), the error bounds were now calculated based
on the local noise levels plus an additional error of 0.05 nm.
This additional error was obtained by analyzing the
obtained distance variations due to an assumed uncertainty
0.15 in the main and side-chain order parameters. The
obtained structures were further refined in explicit solvent
following the protocol by Linge et al., 2003 (80) (see also
Subheadings 3.7.1–3.7.4).
26. For the simulation of NOESY spectra, make sure that the
corresponding parameters are set correctly. Especially give
the correct mixing time, repetition time and
1
H-frequency
(see also Subheading 3.8).
In the last sections, we have seen that automated protein structure
determination in solution at least for smaller well-behaved
proteins is a feasible task. As a consequence, computer-automated
determination of these structures will continue to grow in impor-
tance. However, for difficult test cases such as large biopolymers,
aid from human experts is still required. Typically, in macromo-
lecular NMR, the information content of the NMR spectra alone
is often not sufficient for a complete three-dimensional structure
determination. Signal-to-noise ratios are necessarily limited
due to the limited solubility of most biopolymers. Furthermore,
superpositions of resonance lines often lead to interpretational
ambiguities. Therefore, it is still of importance that any computer
program used for automated structure determination permits
intervention at any step in the analysis to allow the inclusion of
structural and spectroscopic information from other sources and
5. Conclusion

123
Automated Protein NMR Structure Determination in Solution
to supervise various aspects of the validation process. Especially
for the structure improvement by the inclusion of data from
other sources, we have recently developed the AUREMOL-ISIC
(81) algorithm that allows the reliable combination of NMR and
X-ray data.
In addition, the constant development of new experimental
multidimensional NMR techniques requires that new strategies
for automated analysis need to be implemented in existing com-
puter programs. Within this chapter, we have summarized some
of the main points required for automated protein structure
determination in solution. It is clear, however, that due to space
limitations not all possible strategies could be discussed.
Acknowledgments
This work was supported by the Bavarian Genomic Network
BayGene (W. G.) and the European Union (H. R. K.).
References
1. Westbrook, J., Feng, Z., Chen, L., Yang, H.,
Berman, H. M. (2003) The Protein Data Bank
and Structural Genomics. Nucleic Acids Res.
31, 489–491.
2. Gronwald, W., Kalbitzer, H. R. (2004)
Automated Structure Determination of
Proteins by NMR Spectroscopy. Prog. NMR
Spectrosc. 44, 33–96.
3. Huang, Y. P., Moseley, H. N., Baran, M. C.,
Arrowsmith, C., Powers, R., Tejero, R.,
Szyperski, T., Montelione, G. T. (2005) An
Integrated Platform for Automated Analysis of
Protein NMR Structures. Methods Enzymol.
394, 111–141.
4. Güntert, P. (2009) Automated Structure Deter-
mination from NMR Spectra. Eur. Biophys.
J. 38, 129–143.
5. Williamson, M. P., Craven, C. J. (2009)
Automated Protein Structure Calculation from
NMR Data. J. Biomol. NMR 43, 131–143.
6. Kupce, E., Freeman, R. (2004) Projection-
Reconstruction Technique for Speeding Up
Multidimensional NMR Spectroscopy. J. Am.
Chem. Soc. 126, 6429–6440.
7. Hiller, S., Wasmer, S., Wider, G., Wüthrich, K.
(2007) Sequence-Specific Resonance
Assignment of Soluble Nonglobular Proteins
by 7D APSY-NMR Spectroscopy. J. Am.
Chem. Soc. 129, 10823–10828.
8. Savchenko, A., Yee, A., Khachatryan, A.,
Skarina, T., Evdokimova, E., Pavolva, M.,
Semesi, A., Northey, J., Beasley, S., Lan, N.,
Das, R., Gerstein, M., Arrowsmith, C. H.,
Edwards, A. M. (2003) Strategies for Structural
Proteonomics of Prokaryotes: Quantifying the
Advantages of Studying Orthologous Proteins
and of Using Both NMR and X-Ray Crys-
tallography Approaches. Proteins 50, 392–399.
9. Graslund, S., Nordlund, P., Weigelt, J.,
Hallberg, B. M., Bray, J., Gileadi, O., Knapp,
S., Oppermann, U., Arrowsmith, C., Hui, R.,
Ming, J., dhe-Paganon, S., Park, H. W.,
Savchenko, A., Yee, A., Edwards, A.,
Vincentelli, R., Cambillau, C., Kim, R., Kim,
S. H., Rao, Z., Shi, Y., Terwilliger, T. C., Kim,
C. Y., Hung, L. W., Waldo, G. S., Peleg, Y.,
Albeck, S., Unger, T., Dym, O., Prilusky, J.,
Sussman, J. L., Stevens, R. C., Lesley, S. A.,
Wilson, I. A., Joachimiak, A., Collart, F.,
Dementieva, I., Donnelly, M. I., Eschenfeldt,
W. H., Kim, Y., Stols, L., Wu, R., Zhou, M.,
Burley, S. K., Emtage, J. S., Sauder, J. M.,
Thompson, D., Bain, K., Luz, J., Gheyi, T.,
Zhang, F., Atwell, S., Almo, S. C., Bonanno, J.
B., Fiser, A., Swaminathan, S., Studier, F. W.,
Chance, M. R., Sali, A., Acton, T. B., Xiao, R.,
Zhao, L., Ma, L. C., Hunt, J. F., Tong, L.,
Cunningham, K., Inouye, M., Anderson, S.,
Janjua, H., Shastry, R., Ho, C. K., Wang, D.,
124 Gronwald and Kalbitzer
Wang, H., Jiang, M., Montelione, G. T.,
Stuart, D. I., Owens, R. J., Daenke, S., Schutz,
A., Heinemann, U., Yokoyama, S., Bussow, K.,
Gunsalus, K. C. (2008) Protein Produc tion
and Purification. Nat. Methods 5, 135–146.
10. Wood, M. J., Komives, E. A. (1999) Production
of Large Quantities of Isotopically Labeled
Protein in Pichia pastoris by Fermentation.
J. Biomol. NMR 13, 149–159.
11. Nathans, D., Notani, G., Schwartz, J. H.,
Zinder, N. D. (1962) Biosynthesis of the Coat
Protein of Coliphage f2 by E. coli Extracts. Proc.
Natl. Acad. Sci. U. S. A. 48, 1424–1431.
12. Edwards, A. M., Arrowsmith, C. H., Christendat,
D., Dharamsi, A., Friesen, J. D., Greenblatt,
J. F., Vedadi, M. (2000) Protein Production:
Feeding the Crystallographers and NMR spec-
troscopists. Nat. Struc. Biol. 7, 970–972.
13. Noren, C. J., Anthony-Cahill, S. J., Griffith,
M. C., Schultz, P. G. (1989) A General
Method for Site-Specific Incorporation of
Unnatural Amino Acids into Proteins. Science
244, 182–189.
14. Kainosho, M., Torizawa, T., Iwashita, Y.,
Terauchi, T., Mei, O. A., Guntert, P. (2006)
Optimal Isotope Labelling for NMR Protein
Structure Determinations. Nature 440 (7080),
52–57.
15. Sattler, M., Schleucher, J., Griesinger, C.
(1999) Heteronuclear Multidimensional NMR
Experiments for the Structure Determina-
tion of Proteins in Solution Employing Pulsed
Field Gradients. Prog. NMR Spectrosc. 34,
93–158.
16. Cavanagh, J., Fairbrother, W. J., Palmer III, A.
G., Rance, M., Skelton, N. J. (2007) Protein
NMR Spectroscopy. Academic Press: New York.
17. Wishart, D. S., Case, D. A. (2003) Use of
Chemical Shifts in Macromolecular Stru-
cture Determination. Methods Enzymol. 338,
3–34.
18. Szyperski, T., Banecki, B., Glaser, R. W. (1998)
Sequential Resonance Assignment of Medium-
Sized
15
N/
13
C-Labeled Proteins with Projected
4D Triple Resonance Experiments. J. Biomol.
NMR 11, 387–405.
19. Xia, Y., Arrowsmith, C. H., Szyperski, T.
(2002) Novel Projected 4D Triple Resonance
Experiments for Polypeptide Backbone
Chemical Shift Assignment. J. Biomol. NMR
24, 41–50.
20. Szyperski, T., Yeh, D. C., Sukumaran, D. K.,
Moseley, H. N., Montelione, G. T. (2002)
Reduced-Dimensionality NMR Spectroscopy
for High-Throughput Protein Resonance
Assignment. Proc. Natl. Acad. Sci. U. S. A 99,
8009–8014.
21. Schubert, M., Smalla, M., Schmieder, P.,
Oschkinat, H. (1999) MUSIC in Triple-Reso-
nance Experiments: Amino Acid Type-Selective
1
H-
15
N Correlations. J. Magn. Reson. 141, 34–43.
22. Schubert, M., Oschkinat, H., Schmieder, P.
(2001) MUSIC, Selective Pulses, and Tuned
Delays: Amino Acid Type-Selective (1)H-(15)
N Correlations, II. J. Magn Reson. 148,
61–72.
23. Schubert, M., Oschkinat, H., Schmieder, P.
(2001) MUSIC and Aromatic Residues: Amino
Acid Type-Selective (1)H-(15)N Correlations,
III. J. Magn. Reson. 153, 186–192.
24. Schmieder, P., Leidert, M., Kelly, M.,
Oschkinat, H. (1998) Multiplicity-Selective
Coherence Transfer Steps for the Design of
Amino Acid-Selective Experiments-A Triple-
Resonance Experiment Selective for Asn and
Gln. J. Magn. Reson. 131, 199–202.
25. Lopez-Mendez, B., Güntert, P. (2006)
Automated Protein Structure Determination
from NMR Spectra. J. Am. Chem. Soc. 128,
13112–13122.
26. Liu, G., Shen, Y., Atreya, H. S., Parish, D.,
Shao, Y., Sukumaran, D. K., Xiao, R., Yee, A.,
Lemak, A., Bhattacharya, A., Acton, T. A.,
Arrowsmith, C. H., Montelione, G. T.,
Szyperski, T. (2005) NMR Data Collection
and Analysis Protocol for High-Throughput
Protein Structure Determination. Proc. Natl.
Acad. Sci. U. S. A. 102, 10487–10492.
27. Möglich, A., Weinfurtner, D., Maurer, T.,
Gronwald, W., Kalbitzer, H. R. (2005) A
Restraint Molecular Dynamics and Simulated
Annealing Approach for Protein Homology
Modeling Utilizing Mean Angles. BMC
Bioinformatics 6, 91.
28. Möglich, A., Weinfurtner, D., Gronwald, W.,
Maurer, T., Kalbitzer, H. R. (2005) PERMOL:
Restraint-Based Protein Homology Modeling
Using DYANA or CNS. Bioinformatics 21,
2110–2111.
29. Güntert, P. (2004) Automated NMR Structure
Calculation with CYANA. Methods Mol. Biol.
278, 353–378.
30. Brünger, A. T., Adams, P. D., Clore, G. M.,
DeLano, W. L., Gros, P., Grossekunstleve, R.
W., Jiang, J.-S., Kuszewski, J., Nilges, M.,
Pannu, N. S., Read, R. J., Rice, L. M.,
Simonson, T., Warren, G. L. (1998) Crys-
tallography & NMR System: A New Software
Suite for Macromolecular Structure Deter-
mination. Acta Cryst. D 54, 905–921.
31. Schwieters, C. D., Kuszewski, J., Tjandra, N.
L., Clore, G. M. (2003) The Xplor-NIH NMR
Molecular Structure Determination Package.
J. Magn. Reson. 160, 65–73.
125
Automated Protein NMR Structure Determination in Solution
32. Zolani, Z., Macura, S., Markley, J. L. (1989)
Spline Method for Correcting Baseline
Distortions in Two-Dimensional NMR
Spectra. J. Magn. Reson. 82, 496–504.
33. Barsukov, I. L., Arseniev, A. S. (1987) Base-
Plane Correction in 2D NMR. J. Magn. Reson.
73, 148–149.
34. Saffrich, R., Beneicke, W., Neidig, K.-P.,
Kalbitzer, H. R. (1993) Baseline Correction in
n-Dimensional NMR Spectra by Sectionally
Linear Interpolation. J. Magn. Reson. B 101,
304–308.
35. Dietrich, W., Rüdel, C. H., Neumann, M.
(1991) Fast and Precise Automatic Baseline
Correction of One- and Two-Dimensional
NMR Spectra. J. Magn. Reson. 91, 1–11.
36. Golotvin, S., Williams, A. (2000) Improved
Baseline Recognition and Modeling of FT
NMR Spectra. J. Magn. Reson. 146, 122–125.
37. Güntert, P., Wüthrich, K. (1992) FLATT-A
New Procedure for High-Quality Baseline
Correction of Multidimensional NMR Spectra.
J. Magn. Reson. 96, 403–407.
38. Stadlthanner, K., Tome, A. M., Theis, F. J.,
Gronwald, W., Kalbitzer, H. R., Lang, E. W.
(2003) Blind Source Separation of Water
Artefacts in NMR Spectra Using a Matrix
Pencil. Proc. ICA 2003, 167–172.
39. Malloni, W.M., De Sanctis, S., Tome, A.M.,
Lang, E.W., Munte, C.E., Neidig, K.P.,
Kalbitzer, H. R. (2010) Automated solvent artifact
removal and base plane correction of multidi-
mensional NMR protein spectra by AUREMOL-
SSA. J. Biomol. NMR, 47, 101–111.
40. Live, D. H., Davis, D. G., Agosta, W. C.,
Cowburn, D. (1984) Long Range Hydrogen
Bond Mediated Effects in Peptides: 15N NMR
Study of Gramicidin S in Water and Organic
Solvents. J. Am. Chem. Soc. 106, 1939–1941.
41. Maurer, T., Kalbitzer, H. R. (1996) Indirect
Referencing of 31P and 19 F NMR Spectra.
J. Magn. Reson. B 113, 177–178.
42. Markley, J. L., Bax, A., Arata, Y., Hilbers, C.
W., Kaptein, R., Sykes, B. D., Wright, P. E.,
Wüthrich, K. (1998) Recommendations for
the Presentation of NMR Structures of
Proteins and Nucleic Acids. Pure & Appl.
Chem. 70, 117–142.
43. Neidig, K.-P., Bodenmüller, H., Kalbitzer, H. R.
(1984) Computer Aided Evaluation of Two-
Dimensional NMR Spectra of Proteins.
Biochem. Biophys. Res. Comm. 125, 1143–1150.
44. Glaser, S., Kalbitzer, H. R. (1987) Automated
Recognition and Assessment of Cross Peaks in
Two-Dimensional NMR Spectra of Macro-
molecules. J. Magn. Reson. 74, 450–463.
45. Pfändler, P., Bodenhausen, G. (1988) Analysis of
Multiplets in Two-Dimensional NMR Spectra
by Topological Classification: Applications to
Vinblastine and Cyclosporin A. Magn. Reson.
Chem. 26, 888–894.
46. Novic, M., Eggenberger, U., Bodenhausen, G.
(1988) Similarities Between Self-Convolution
and Symmetry Mapping of Multiplets in Two-
Dimensional NMR Spectra. J. Magn. Reson.
77, 394–400.
47. Stoven, V., Mikou, A., Piveteau, D., Guitettet,
E., Lallemand, J.-Y. (1989) PARIS, a Program
for Automatic Recognition and Integration of
2D NMR Signals. J. Magn. Reson. 82, 163–168.
48. Eccles, C., Guntert, P., Billeter, M., Wuthrich,
K. (1991) Efficient analysis of protein 2D
NMR spectra using the software package
EASY. J. Biomol. NMR 1, 111–130.
49. Koradi, R., Billeter, M., Engeli, M., Guntert,
P., Wuthrich, K. (1998) Automated Peak
Picking and Peak Integration in Macromolecular
NMR Spectra Using AUTOPSY. J. Magn.
Reson. 135, 288–297.
50. Neidig, K.-P., Geyer, M., Görler, A., Antz, C.,
Saffrich, R., Beneicke, W., Kalbitzer, H. R.
(1995) AURELIA, a Program for Computer-
Aided Analysis of Multidimensional NMR
Spectra. J. Biomol. NMR 6, 255–270.
51. Neidig, K.-P., Kalbitzer, H. R. (1990)
Improved Representation of Two-Dimensional
NMR Spectra by Local Rescaling. J. Magn.
Reson. 88, 155–160.
52. Antz, C., Neidig, K.-P., Kalbitzer, H. R. (1995)
A General Bayesian Method for an Automated
Signal Class Recognition in 2D NMR Spectra
Combined with a Multivariate Discriminant
Analysis. J. Biomol. NMR 5, 287–296.
53. Schulte, A. C., Gorler, A., Antz, C., Neidig, K.
P., Kalbitzer, H. R. (1997) Use of Global
Symmetries in Automated Signal Class
Recognition by a Bayesian Method. J. Magn.
Reson. 129, 165–172.
54. Geyer, M., Neidig, K.-P., Kalbitzer, H. R.
(1995) Automated Peak Integration in
Multidimensional NMR Spectra by an
Optimized Iterative Segmentation Procedure.
J. Magn. Reson. B 109, 31–38.
55. Denk, W., Baumann, R., Wagner, G. (1986)
Quantitative Evaluation of Cross-Peak
Intensities by Projection of Two-Dimensional
NOE Spectra on a Linear Space Spanned by a
Set of Reference Resonance Lines. J. Magn.
Reson. 67, 386–390.
56. Kobayashi, N., Iwahara, J., Koshiba, S.,
Tomizawa, T., Tochio, N., Güntert, P.,
Kigawa, T., Yokoyama, S. (2007) KUJARA, a
126 Gronwald and Kalbitzer
Package of Integrated Modules for Systematic
and Interactive Analysis of NMR Data Directed
to High-Throughput NMR Structure Studies.
J. Biomol. NMR 39, 31–52.
57. Linge, J. P.; Habeck, M.; Rieping, W.; Nilges,
M. (2003) ARIA: Automated NOE Assignment
and NMR Structure Calculation. Bioinformatics
19, 315–316.
58. Herrmann, T., Güntert, P., Wüthrich, K.
(2002) Protein NMR Structure Determination
with Automated NOE-Identification in the
NOESY Spectra Using the New Software
ATNOS. J. Biomol. NMR 24, 171–189.
59. Gronwald, W., Moussa, S., Elsner, R., Jung,
A., Ganslmeier, B., Trenner, J., Kremer,
W., Neidig, K. P., Kalbitzer, H. R. (2002)
Automated Assignment of NOESY NMR
Spectra Using a Knowledge Based Method
(KNOWNOE). J. Biomol. NMR 23, 271–287.
60. Geyer, M., Herrmann, C., Wohlgemuth, S.,
Wittinghofer, A., Kalbitzer, H. R. (1997)
Structure of the Ras-Binding Domain of
RalGEF and Implications for Ras Binding and
Signalling. Nat. Struc. Biol. 4, 694–699.
61. Gronwald, W., Brunner, K., Kirchhöfer, R.,
Trenner, J., Neidig, K.-P., Kalbitzer, H. R.
(2007) AUREMOL-RFAC-3D, Combination
of R-Factors and Their Use for Automated
Quality Assessment of Protein Structures.
J. Biomol. NMR 37, 15–30.
62. Gorler, A., Gronwald, W., Neidig, K. P.,
Kalbitzer, H. R. (1999) Computer Assisted
Assignment of 13C or 15N Edited 3D-NOESY-
HSQC Spectra Using Back Calculated and
Experimental Spectra. J. Magn. Reson. 137,
39–45.
63. Görler, A., Kalbitzer, H. R. (1997) Relax, a
Flexible Program for the Back Calculation of
NOESY Spectra Based on Complete-
Relaxation-Matrix Formalism. J. Magn. Reson.
124, 177–188.
64. Ried, A., Gronwald, W., Trenner, J. M.,
Brunner, K., Neidig, K.-P., Kalbitzer, H. R.
(2004) Improved Simulation of NOESY
Spectra by RELAX-JT2 Including Effects of
J-Coupling, Transverse Relaxation and Chemical
Shift Anisotropy. J. Biomol. NMR 30, 121–131.
65. Keepers, J. W., James, T. L. (1984) A Theo-
retical Study of Distance Determination from
NMR. Two-Dimensional Nuclear Overhauser
Effect Spectra. J. Magn. Reson. 57, 404–426.
66. Boelens, R., Koning, M. G., van der Marel, G. A.,
van Boom, J. H., Kaptein, R. (1989) Iterative
Procedure for Structure Determination from
Proton-Proton NOEs Using a Full Relaxation
Matrix Approach. Application to a DNA
Octamer. J. Magn. Reson. 82, 290–380.
67. Borgias, B. A., James, T. L. (1990)
MARDIGRAS-A Procedure for Matrix
Analysis of Relaxation for Discerning Geometry
of an Aqueous Structure. J. Magn. Reson. 87,
475–487.
68. Post, C. B., Meadows, R. P., Gorenstein, D.
G. (1990) On the Evaluation of Interproton
Distances for Three-Dimensional Structure
Determination by NMR Using a Relaxation
Rate Matrix Analysis. J. Am. Chem. Soc. 112,
6796–6803.
69. van de Ven, F. J. M., Blommers, M. J. J.,
Schouten, R. E., Hilbers, C. W. (1991)
Calculation of Interproton Distances from
NOE Intensities. A Relaxation Matrix
Approach Without Requirement of a Molecular
Model. J. Magn. Reson. 94, 140–151.
70. Madrid, M., Llinas, E., Llinas, M. (1991)
Model-Independent Refinement of Inter-
proton Distances Generated from
1
H NMR
Overhauser Intensities. J. Magn. Reson. 93,
329–346.
71. Kim, S.-G., Reid, B. R. (1992) Automated
NMR Structure Refinement via NOE Peak
Volumes. Application to a Dodecamer DNA
Duplex. J. Magn. Reson. 100, 382–390.
72. Moseley, H. N. B., Curto, E. V., Krishna, N. R.
(1995) Complete Relaxation and Conforma-
tional Exchange Matrix (CORCEMA) Analysis
of NOESY Spectra of Interacting Systems;
Two-Dimensional Trans ferred NOESY. J. Magn.
Reson. B 108, 243–261.
73. Ernst, R. R., Bodenhausen, G., Wokaun, A.
(1987) Principles of Nuclear Magnetic Reso-
nance in One and Two Dimensions. Clarendon
Press: Oxford.
74. Jeener, J., Meier, B. H., Bachmann, P., Ernst,
R. R. (1979) Investigation of Exchange Pro-
cesses by Two-Dimensional NMR Spectro-
scopy. J. Chem. Phys. 71, 4546–4553.
75. Macura, S., Ernst, R. R. (1980) Elucidation of
Cross Relaxation in Liquids by Two-
Dimensional N.M.R. Spectroscopy. Mol. Phys.
41, 95–117.
76. Boelens, R., Koning, M. G., Kaptein, R. (1988)
Determination of Biomolecular Structures from
Protein-Protein NOE’s Using a Relaxa tion
Matrix Approach. J. Mol. Struct. 173, 299–311.
77. Gronwald, W., Kirchhofer, R., Gorler, A.,
Kremer, W., Ganslmeier, B., Neidig, K. P.,
Kalbitzer, H. R. (2000) RFAC, a Program for
Automated NMR R-Factor Estimation.
J. Biomol. NMR 17, 137–151.
78. Baskaran, K., Kirchhofer, R., Huber, F.,
Trenner, J., Brunner, K., Gronwald, W.,
Neidig, K. P., Kalbitzer, H. R. (2009) Chemical
Shift Optimization in Multidimensional NMR
127
Automated Protein NMR Structure Determination in Solution
Spectra by AUREMOL-SHIFTOPT. J. Biomol.
NMR 43, 197–210.
79. Linge, J. P., O’Donoghue, S. I., Nilges, M.
(2001) Automated Assignment of Ambiguous
Nuclear Overhauser Effects with ARIA.
Methods Enzymol. 339, 71–90.
80. Linge, J. P., Williams, M. A., Spronk, C. A. E. M.,
Bonvin, A. M. J. J., Nilges, M. (2003)
Refinement of Protein Structures in Explicit
Solvent. Proteins 50, 496–506.
81. Brunner, K., Gronwald, W., Trenner, J. M.,
Neidig, K.-P., Kalbitzer, H. R. (2006) A gen-
eral Method for the Unbiased Improvement
of Solution NMR Structures by the Use of
Related X-Ray Data, the AUREMOL-ISIC
Algorithm. BMC Struc. Biol. 6, 14.

129
Chapter 8
Computational Tools in Protein Crystallography
Deepti Jain and Valerie Lamour
Abstract
Protein crystallography emerged in the early 1970s and is, to this day, one of the most powerful techniques
for the analysis of enzyme mechanisms and macromolecular interactions at the atomic level. It is also an
extremely powerful tool for drug design. This field has evolved together with developments in computer
science and molecular biology, allowing faster three-dimensional structure determination of complex bio-
logical assemblies. In recent times, structural genomics initiatives have pushed the development of meth-
ods to further speed up this process. The algorithms initially defined in the last decade for structure
determination are now more and more elaborate, but the computational tools have evolved toward simpler
and more user-friendly packages and web interfaces. We present here a modest overview of the popular
software packages that have been developed for solving protein structures, and give a few guidelines and
examples for structure determination using the two most popular methods, molecular replacement and
multiple anomalous dispersion.
Key words: X-ray crystallography, Protein structure determination, Crystallography software,
Molecular replacement, Multi-wavelength anomalous dispersion
The correct three-dimensional (3D) arrangement of constituent
atoms is central to the proper functioning of a majority of biologi-
cal molecules. This fact is especially true of proteins, and hence, it
is important to determine the structures of macromolecules and
their assemblies to understand their function in great detail. X-ray
crystallography is an experimental science that has benefited tre-
mendously from all the software and hardware developments dur-
ing the past three decades. Structure determination through X-ray
crystallography is becoming more and more automated through
the development of user-friendly interfaces and new crystallogra-
phy packages. These developments have made the method more
accessible to the biochemist with little theoretical background in
1. Introduction
David Fenyö (ed.), Computational Biology, Methods in Molecular Biology, vol. 673,
DOI 10.1007/978-1-60761-842-3_8, © Springer Science+Business Media, LLC 2010
130 Jain and Lamour
crystallography. Often behind every structure lies months or more
commonly years of bench work to clone, express, and purify mil-
ligram quantities of protein required to produce diffracting crystals,
a prerequisite for any structural work using X-ray crystallography.
Useful information regarding this aspect of the method can be
found in numerous reviews that summarize decades of research in
structural biology, spanning simple proteins to high-molecular
weight biological macromolecular assemblies (1–3). The present
chapter will focus on the major methods and tools available to
determine the 3D structure of a macromolecule once diffracting
crystals have been prepared. It provides a concise reference to a
beginner in protein crystallography.
Computational tools in the form of program packages are con-
stantly evolving to feed the need for faster and more convenient
structure determination, especially driven by structural genomics
initiatives. We provide here a brief overview of the most common
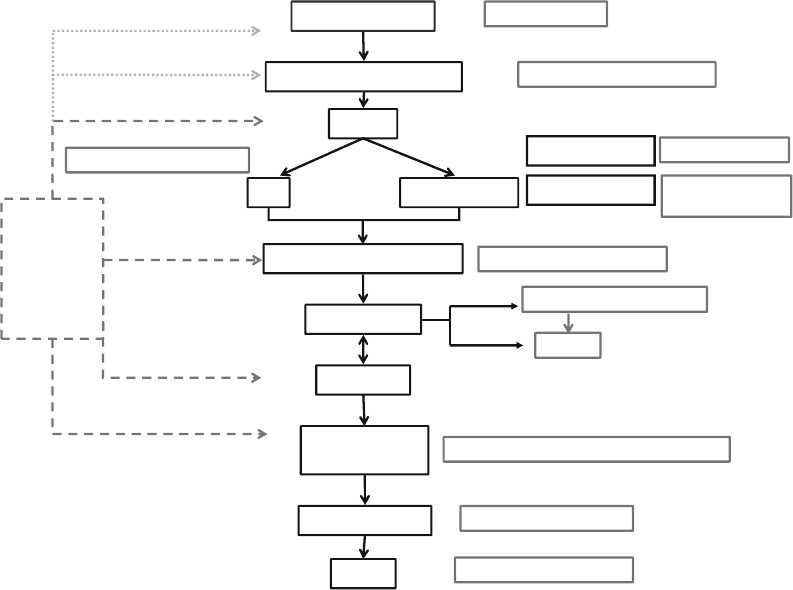
and most widely used crystallographic packages (see Fig. 1) and
guidelines for structure determination using the favored phasing
Data collection
Data processing and scaling
HKL2000, Mosflm , XDS, d*trek
Refinement
Integrated
packages :
CNS
CCP4
Phenix
MRBump
AutoRickshaw
HKL3000
Density modification
Model building
Phasing
Model validation
PDB submission
O, Coot
LIGPLOT, CNS, Grasp
MAD, SAD, MIR
MR
Structure analysis
Dino, Rasmol, Pymol
Procheck, Sfcheck, WHAT_CHECK, Molprobity
Figures
Strategy, BEST
SnB, SHELX, SOLVE
Amore, Molrep, Phaser, EPMR
SHARP, MLPHARE,
SOLVE
DM, SOLOMON, RESOLVE
ARP/wARP, SOLVE/RESOLVE
Automated
Manual
Heavy atom search
Refinement & Phasing
Fig. 1. General scheme representing the different steps to solve a crystallographic structure. A selection of programs is
reported next to each step of X-ray structure determination. Program packages or pipelines (dash line box) cover several
steps of structure determination. HKL3000 also includes data processing in the pipeline.
