Feny? D. (Ed.) Computational Biology
Подождите немного. Документ загружается.


253
Chapter 17
Reverse Engineering Gene Regulatory Networks Related
to Quorum Sensing in the Plant Pathogen Pectobacterium
atrosepticum
Kuang Lin, Dirk Husmeier, Frank Dondelinger, Claus D. Mayer, Hui Liu,
Leighton Prichard, George P.C. Salmond, Ian K. Toth,
and Paul R.J. Birch
Abstract
The objective of the project reported in the present chapter was the reverse engineering of gene regula-
tory networks related to quorum sensing in the plant pathogen Pectobacterium atrosepticum from mic-
orarray gene expression profiles, obtained from the wild-type and eight knockout strains. To this end, we
have applied various recent methods from multivariate statistics and machine learning: graphical Gaussian
models, sparse Bayesian regression, LASSO (least absolute shrinkage and selection operator), Bayesian
networks, and nested effects models. We have investigated the degree of similarity between the predic-
tions obtained with the different approaches, and we have assessed the consistency of the reconstructed
networks in terms of global topological network properties, based on the node degree distribution. The
chapter concludes with a biological evaluation of the predicted network structures.
Key words: Pectobacterium atrosepticum, Quorum sensing, Transposon mutagenesis, Microarrays,
Graphical Gaussian models, Sparse Bayesian regression, LASSO, Bayesian networks, Nested effects
models, Degree distribution, Power law, Gene ontologies
Pectobacterium atrosepticum (Pba), which is a plant pathogen on
potato in temperate regions, synthesizes and secretes large quan-
tities of plant cell wall degrading enzymes that are responsible for
the soft rot disease phenotype, earning it the epithet “brute force”
pathogen. In particular, the “brute force” attack utilizes a population
density-dependent regulatory mechanism called quorum sens-
ing (QS), which controls a wide range of phenotypes in many
1. Introduction
David Fenyö (ed.), Computational Biology, Methods in Molecular Biology, vol. 673,
DOI 10.1007/978-1-60761-842-3_17, © Springer Science+Business Media, LLC 2010

254 Lin et al.
different bacteria. Utilizing the production and secretion of certain
signalling molecules, QS serves as a communication network that
allows bacteria to coordinate their activities based on the local
density of their population. A recent study by Liu et al. (1) pro-
vides the first evidence that Pba uses QS to target host defences
simultaneously with a physical attack on the plant cell wall.
Moreover, Liu et al. (1) demonstrate that a wide range of previ-
ously known and unknown virulence regulators lie within the QS
regulon, revealing it to be the master regulator of virulence. The
objective of the present study is to shed further light on the QS
regulatory mechanism by applying current methods from multi-
variate statistics and machine learning to reconstruct putative
gene regulatory networks from gene expression profiles obtained
from wild-type and various knockout strains.
Mutated bacterial strains were generated via transposon muta-
genesis. Transposons are relatively short pieces of mobile DNA
that can insert into pieces of DNA within a genome. Transposon
mutagenesis is a process that allows transposons to be transferred
to a host organism’s chromosome. This is accomplished by way of
a plasmid from which a transposon is extracted and inserted into
the host chromosome. The insertion can result in the interrup-
tion or modification of the function of an extant gene on the
chromosome, effectively creating a mutant knockout strain. In
the present study, nine mutant Pba strains were generated, where
the following genes were knocked out: expM, hor, hrpL, expI,
expR, aepA, virR, and virS. Additionally, a double mutation event
was induced, where both virR and expI were knocked out. For
further details and an exact specification of the experimental
protocol, see ref. (1).
Wild-type and mutant Pba strains were grown in a nutrient broth to
stationary phase, and then used to inoculate sterilized potato tubers.
At 12 h postinoculation, the bacterial cells were isolated from the
tuber by scraping infected tissue into sterilised water. RNA was iso-
lated by following the protocol described in Liu et al. (1), then
reverse transcribed and cDNA labelled. 60-Mer oligonucleotide
probes were designed to Pba-coding sequences and used, together
with controls, to generate 11K custom arrays with 99.5% genome
coverage (Agilent, Inc., Santa Clara, CA, USA). All microarray
experiments were carried out in triplicate, for each of the eight single
Pba knockout mutants in expM, hor, hrpL, expI, expR, aepA, virR,
and virS, and the double knockout mutant in virR/expI, to obtain
relative gene expression levels with respect to Pba wild-type.
2. Material
2.1. Gene Knockout via
Transposon
Mutagenesis
2.2. Genome-Wide
Transcriptomic
Profiling with
Miroarrays

255
Reverse Engineering Gene Regulatory Networks
All microarray images were visually assessed for quality prior to
feature extraction, whereby standard probe quality control stan-
dards were applied
1
. Features flagged as poor were removed. Box
plots and principal components analysis of whole data sets were used
to assess array to array variation. Any outlying microarrays were
repeated as necessary. The microarray data were preprocessed using
GeneSpring
2
software (version 7.2) and normalized using the
Lowess algorithm (Agilent Technologies Inc.). This nonparametric
normalization technique first fits a nonlinear curve to the plot of the
log-ratios of the two dye intensities Cy5 and Cy3, M = log
2
(Cy5/Cy3),
versus the average log-ratio A = log
2
(Cy3 * Cy5)/2. It then uses the
residuals of the fit as normalized log-ratio values. This method was
first suggested by Yang et al. (2) and has become the standard
method of normalizing two-colour microarrays.
3
To assess differential gene expression in Pba knockout strains with
respect to Pba wildtype, we computed p- values with the empirical
Bayes method proposed in Smyth (3). Recall that all knockout
experiments were carried out in triplicate. The three resulting log-
ratio values for each mutant versus wild-type comparison were tested
against 0 by using the moderated t-test available in the Bioconductor
library LIMMA (4). This test differs from a standard t-test by using
a standard error estimate that is obtained from an empirical Bayesian
analysis. The individual estimate for a single gene is shrunk towards
the average estimate for all genes, which stabilizes the analysis par-
ticularly for small sample sizes, as in our example. This method was
first introduced by Lönnstedt and Speed (5), later generalized and
implemented in Bioconductor
4
by Smyth (3), and it is one of the
most widely used tools for detecting differential gene expression. As
a result of this analysis, we obtain a p- value for each gene, which
indicates whether the corresponding average log-ratio between
mutant and wild-type is significantly different from 0.
It would be difficult to visualize and interpret regulatory networks
involving several thousand genes. Moreover, there would be con-
siderable inference uncertainty, as the likelihood in network space
would be diffuse, with many different networks having very similar
scores. We therefore resorted to a clustering approach as a preliminary
2.3. Preprocessing
of Gene Expression
Profiles
2.4. Assessing
Differential Gene
Expression
2.5. Clustering of Gene
Expression Profiles
1
See further information in ArrayExpress-http://www.ebi.ac.uk/microarray-
as/aer/.
2
http://www.chem.agilent.com/en-US/Products/software/lifesciences-
informatics/genespringgx/Pages/default.aspx.
3
Note that in contrast to most home-spotted cDNA microarrays, Agilent
arrays are not printed by different print tips and thus are not subdivided into
separate subblocks within the array. For this reason, it was not necessary to
use the print-tip lowess algorithm, which applies the same curve fitting tech-
nique separately to each subblock on the array.
4
http://www.bioconductor.org/.
256 Lin et al.
complexity reduction step, and then inferred regulatory interactions
among about 100 inferred clusters and nine key target regulatory
genes. In order to infer biologically plausible clusters, we combined
the outputs from several clustering algorithms based on a biologi-
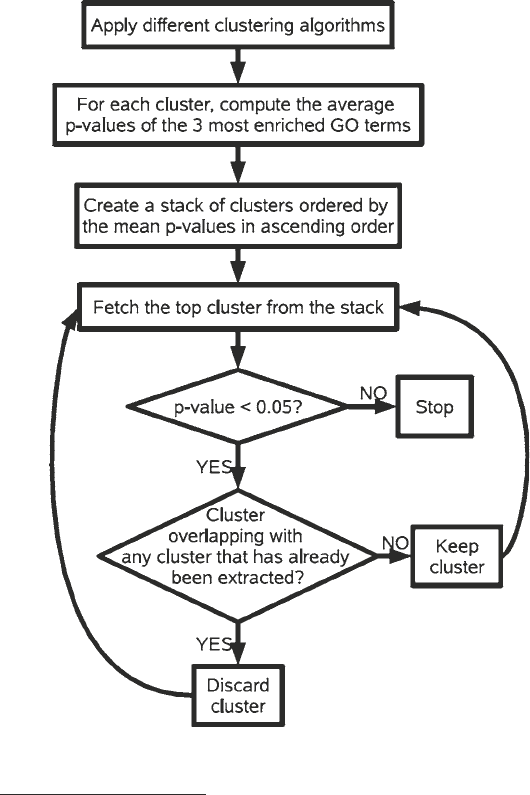
cal scoring scheme using gene ontologies. The basic idea is depicted
in the flow chart of Fig. 1. We applied five clustering algorithms:
K-means and hierarchical agglomerative average linkage clustering
(6), using both Euclidean and correlation distances, as well as mix-
tures of factor analyzers inferred with variational Bayesian
Expectation Maximization (7). We assessed the biological plausibil-
ity of the inferred clusters by testing for significantly enriched GO
(Gene Ontology) terms. We collected GO annotations for both
Pba genes and close homologues from the EBI
5
and ASAP
6
data-
bases. In total, 18,996 GO terms were assigned to the 3,616 genes
Fig. 1 Flow chart of the algorithm used for clustering gene expression profiles.
5
http://www.ebi.ac.uk/GOA/proteomes.html.
6
https://asap.ahabs.wisc.edu/asap/logon.php.
257
Reverse Engineering Gene Regulatory Networks
in our set. We computed the significance of GO term enrichment
in the clusters using the program Ontologizer
7
with the default
options. We applied a standard 5% threshold cutoff on the
Bonferroni-corrected p- values, and considered a cluster to be bio-
logically plausible when the p- value of a GO term enrichment was
smaller than 5%. We then combined clusters from different cluster-
ing methods by the application of the algorithm depicted in Fig. 1,
resulting in 110 clusters. The composition of these clusters, as well
as further details of the clustering scheme, can be obtained from
the supplementary material
8
.
A simple method for reconstructing gene regulatory networks,
proposed by Butte and Kohane (8) and termed “relevance net-
works,” is based on the following procedure. First, compute all
pairwise similarity scores between gene expression profiles.
Standard measures of similarity that are commonly used are the
Pearson correlation or the mutual information. Next, apply a ran-
domization test to test for significant deviation from zero. Finally,
connect all nodes by edges whose pairwise similarity scores are
significantly greater than zero. The method is computationally
cheap and easy to apply. However, its main disadvantage is that it
is intrinsically impossible to distinguish between direct and indi-
rect interactions. If two genes are regulated by a set of common
regulators, their gene expression profiles tend to be similar. The
relevance network approach will therefore tend to infer an edge
between these genes even if there is no direct interaction between
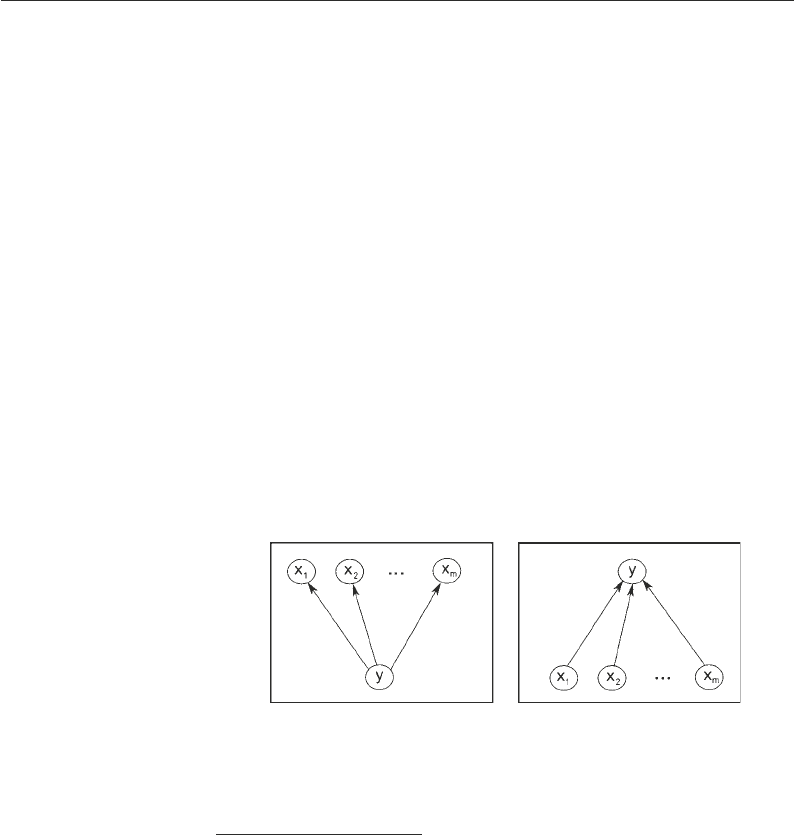
them. For instance, in the scenario depicted in the left panel of
Fig. 2, where a set of m genes x
1
, x
2
, …, x
m
are regulated by the
3. Methods
Fig. 2. Schematic of the approach of partial correlation (left) and sparse regression (right ).
Left : Conditional on y, the gene expression profiles x
1
, x
2
, …, x
m
are independent, and the
partial correlation coefficients will be small. Right : The approach of sparse regression aims
to find a minimal set of predictors x
1
,x
2
, …, x
m
to explain gene expression profile y.
7
http://www.charite.de/ch/medgen/ontologizer/commandline/
Ontologizer.jar.
8
http://www.bioss.ac.uk.testweb.bioss.sari.ac.uk/staff/dirk/
Supplements/FF842/.

258 Lin et al.
common regulator y, the approach of relevance networks is prone
to inferring spurious edges between the genes x
1
, x
2
, …, x
m
; see
Werhli et al. (9) for an empirical corroboration.
In the following subsections, we will briefly review various
more sophisticated methods that aim to distinguish direct inter-
actions from indirect ones, and which also give some indication
about the putative direction of the regulatory interactions. We
first assume that we have complete observability, i.e. that the gene
expression profiles provide a good indication of the correspond-
ing protein activities. We review four approaches aiming to infer
gene regulatory networks from expression profiles: Graphical
Gaussian models, LASSO, sparse Bayesian regression, and
Bayesian networks. We conclude this section with a review of
nested effects models, which aim to infer interactions among reg-
ulatory genes that are themselves subject to post-transcriptional
modification. This approach allows regulatory gene interactions
to be inferred when the data are incomplete, i.e., when the rele-
vant changes at the protein level are not indicated by changes at
the gene expression level.
Graphical Gaussian models (GGMs) are undirected probabilistic
graphical models that allow the identification of conditional inde-
pendence relations among the nodes under the assumption of a
multivariate Gaussian distribution of the data. The inference of
GGMs is based on a (stable) estimation of the covariance matrix
of this distribution. The element C
ik
of the covariance matrix C is
proportional to the correlation coefficient between nodes X
i
and
X
k
. A high correlation coefficient between two nodes may indi-
cate a direct interaction, an indirect interaction, or a joint regula-
tion by a common (possibly unknown) factor. However, only the
direct interactions are of interest to the construction of a regula-
tory network. The strengths of these direct interactions are mea-
sured by the partial correlation coefficient r
ik
, which describes the
correlation between nodes X
i
and X
k
conditional on all the other
nodes in the network. From the theory of normal distributions, it
is known that the matrix of partial correlation coefficients r
ik
is
related to the inverse of the covariance matrix C, C
−1
(with ele-
ments
1
ik
C
−
(10).
−
−−
ρ =−
1
11
.
ik
ik
ii kk
C
CC
(1)
To infer a GGM, one typically employs the following procedure.
From the given data, the empirical covariance matrix is computed,
inverted, and the partial correlations r
ik
are computed from (1).
The distribution of | r
ik
| is inspected, and edges (i, k) corre-
sponding to significantly small values of |r
ik
| are removed from
3.1. Graphical
Gaussian Models
259
Reverse Engineering Gene Regulatory Networks
the graph. The critical step in the application of this procedure is
the stable estimation of the covariance matrix and its inverse.
Note that the covariance matrix is only nonsingular if the number
of observations exceeds the number of nodes in the network. This
condition is not satisfied for many real applications in systems
biology. In order to learn a GGM from a data set in such a sce-
nario, Schäfer and Strimmer (11) explored various stabilization
methods, based on the Moore–Penrose pseudo inverse and bag-
ging. In the present work, we apply an alternative regularization
approach based on shrinkage, which Schäfer and Strimmer (11)
found to be superior to their earlier schemes. The idea is to add a
weighted nonsingular regularization matrix, e.g., the unity matrix,
to the covariance matrix so as to guarantee its nonsingularity. The
optimal weight parameter is estimated based on the Ledoit Wolf
lemma from statistical decision theory so as to minimize the expected
deviation of the regularized covariance matrix from the (unknown)
true covariance matrix. The method of GGMs, which are undi-
rected graphs, can be extended to infer putative directions of
causal interactions, as proposed in Opgen-Rhein and Strimmer
(12). This scheme is based on the computation of the standard-
ized partial variance, which is the proportion of the variance that
remains if the influence of all other variables is taken into account.
All significant edges in the GGM network are directed in such a
fashion that the direction of the arrow points from the node with
the larger standardized partial variance (the more exogeneous node)
to the node with the smaller standardized partial variance (the
more endogeneous node), provided the ratio of the two partial
variances is significantly different from 1. For further details, see
ref. (12).
The approach discussed in the previous subsection aims to predict
interactions between genes based on the partial correlations
between their expression profiles. In the present subsection, we
review an alternative paradigm, which pursues a regression
approach: given the gene expression profile y
g
of some target gene
g, we aim to find a set of regulator genes {r} whose gene expression
profiles {x
r
} are good predictors of gene expression profile y
g
:
=
∑
r
ˆ
,
g gr
r
w xy
(2)
where ŷ
g
is a predictor of y
g
, and the regression parameters w
gr
represent interaction strengths between the target gene g and the
putative regulator genes r. The different concepts are illustrated
in Fig. 2. We denote the vector of interaction strengths as w
g
,
which has w
gr
as its rth component. The mismatch between the
predicted and measured expression profile of target gene g is typi-
cally measured by the L2 norm
3.2. Sparse Regression
and the LASSO

260 Lin et al.
=−
2
ˆ
() ().
g g gg
EYwy w
(3)
Obtaining the optimal interaction parameters ŵ
g
by minimizing
E(w
g
) corresponds to a maximum likelihood estimator under the
assumption of isotropic Gaussian noise. In practice, this approach
is usually susceptible to overfitting, which calls for the application
of some regularization scheme. The standard method of ridge
regression is given by
= +λ
∑
2
argmin ( ) .
g
g
g gr
r
Ew
w
ww
(4)
This can be interpreted in three different ways: (1) maximizing
the penalized likelihood with an L2-norm penalty term and regu-
larization parameter l; (2) constrained maximization of the likeli-
hood under the L2-norm constraint
<∑
2
r gr
wC
, where l is a
Lagrange parameter; (3) Bayesian maximum a posteriori estimate
under a zero-mean Gaussian prior on w
g
with diagonal isotropic
covariance matrix l
−1
I: P(w
g
) =
- 2008 — 2025 «СтудМед»
