Berg J.M., Tymoczko J.L., Stryer L. Biochemistry
Подождите немного. Документ загружается.


The other two domains of the tripartite EF-Tu are distinctive; they mediate interactions with aminoacyl-tRNA and the
ribosome. In all these related enzymes, the change in conformation between the GTP and the GDP forms leads to a
change in interaction partners. A further similarity is the requirement that an additional protein catalyze the exchange of
GTP for GDP; an activated receptor plays the role of EF-Ts for a heterotrimeric G protein, as does Sos for Ras.
29.4.3. The Formation of a Peptide Bond Is Followed by the GTP-Driven Translocation
of tRNAs and mRNA
After the correct aminoacyl-tRNA has been placed in the A site, the transfer of the polypeptide chain from the tRNA in
the P site is a spontaneous process, driven by the formation of the stronger peptide bond in place of the ester linkage.
However, protein synthesis cannot continue without the translocation of the mRNA and the tRNAs within the ribosome.
The mRNA must move by a distance of three nucleotides as the deacylated tRNA moves out of the P site into the E site
on the 30S subunit and the peptidyl-tRNA moves out of the A site into the P site on the 30S subunit. The result is that the
next codon is positioned in the A site for interaction with the incoming aminoacyl-tRNA.
Translocation is mediated by elongation factor G (EF-G, also called translocase). The structure of EF-G is exceptional
in revealing some aspects of its mode of action (Figure 29.29). The structure of EF-G closely resembles that of the
complex between EF-Tu and tRNA. This is an example of molecular mimicry; a protein domain evolved so that it
mimics the shape of a tRNA molecule. This structural similarity, as well as other experimental data, suggests a
mechanism for the translocation process (Figure 29.30). First, EF-G in the GTP form binds to the ribosome, primarily
through the interaction of its EF-Tu-like domain with the 50S subunit. The binding site includes proteins L11 and the L7-
L12 dimer. The tRNA-like domain of EF-G interacts with the 30S subunit. The binding of EF-G to the ribosome in this
manner stimulates the GTPase activity of EF-G. On GTP hydrolysis, EF-G undergoes a conformational change that
forces its arm deeper into the A site on the 30S subunit. To accommodate this domain, the peptidyl-tRNA in the A site
moves to the P site, carrying the mRNA and the deacylated tRNA with it. The ribosome may be prepared for these
rearrangements by the initial binding of EF-G as well. The dissociation of EF-G leaves the ribosome ready to accept the
next aminoacyl-tRNA into the A site.
29.4.4. Protein Synthesis Is Terminated by Release Factors That Read Stop Codons
The final phase of translation is termination. How does the synthesis of a polypeptide chain come to an end when a stop
codon is encountered? Aminoacyl-tRNA does not normally bind to the A site of a ribosome if the codon is UAA, UGA,
or UAG, because normal cells do not contain tRNAs with anticodons complementary to these stop signals. Instead, these
stop codons are recognized by release factors (RFs), which are proteins. One of these release factors, RF1, recognizes
UAA or UAG. A second factor, RF2, recognizes UAA or UGA. A third factor, RF3, another G protein homologous to
EF-Tu, mediates interactions between RF1 or RF2 and the ribosome.
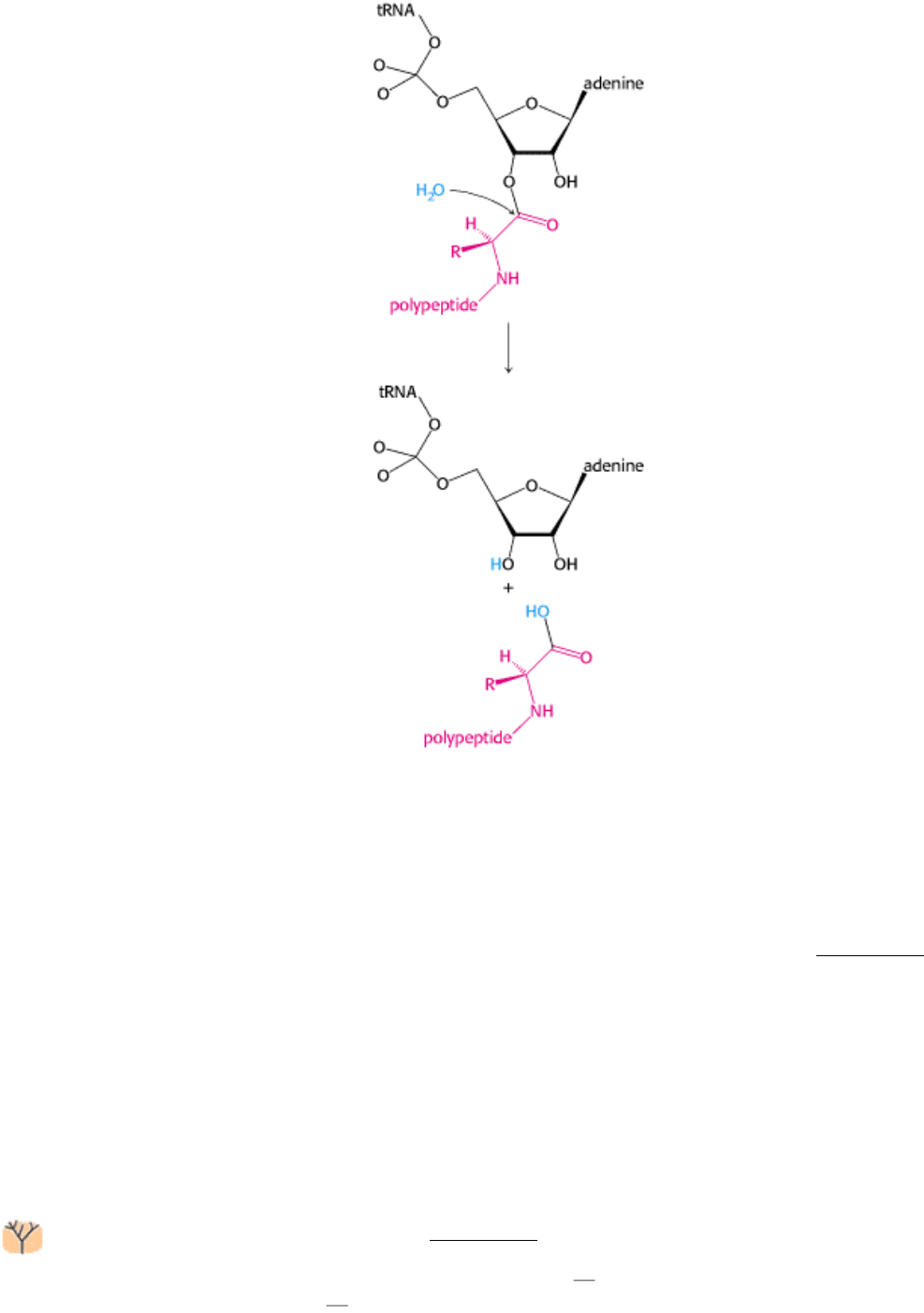
Release factors use a Trojan horse strategy to free the polypeptide chain. One of the most impressive properties of the
ribosome is not that it catalyzes peptide-bond formation; the formation of a peptide bond by the reaction between an
amino group and an ester is a facile chemical reaction. Instead, a more impressive feature crucial to ribosome function is
that the peptidyl-tRNA ester linkage is not broken by premature hydrolysis. The exclusion of water from the peptidyl
transferase center is crucial in preventing such hydrolysis, which would lead to release of the polypeptide chain. The
structure of a prokaryotic release factor has not yet been determined. However, the structure of a eukaryotic release
factor, though probably not truly homologous to its prokaryotic counterpart, reveals the strategy (Figure 29.31).
The structure resembles that of a tRNA by molecular mimicry. The sequence Gly-Gly-Gln, present in both eukaryotes
and prokaryotes, occurs at the end of the structure corresponding to the acceptor stem of a tRNA. This region binds a
water molecule. Disguised as an aminoacyl-tRNA, the release factor may carry this water molecule into the peptidyl
transferase center and, assisted by the catalytic apparatus of the ribosome, promote this water molecule's attack on the
ester linkage, freeing the polypeptide chain. The detached polypeptide leaves the ribosome. Transfer RNA and
messenger RNA remain briefly attached to the 70S ribosome until the entire complex is dissociated in a GTP-dependent
fashion by ribosome release factor (RRF) and EF-G. Ribosome release factor is an essential factor for prokaryotic
translation.
The structure of RRF, too, resembles tRNA (Figure 29.32). However, the known tRNA-mimicking structures of
RRF, EF-G, and the release factors are distinct; they do not appear to have been generated from a common
ancestor. Thus, convergent evolution has provided a similar solution looking sufficiently like a tRNA to interact with
the tRNAbinding sites on the ribosome
to several problems. The effects of divergentevolution are evident in the
protein factors that participate in translation, most notably in the form of the homologous G proteins, EF-Tu, EF-G, IF2,
and RF3.
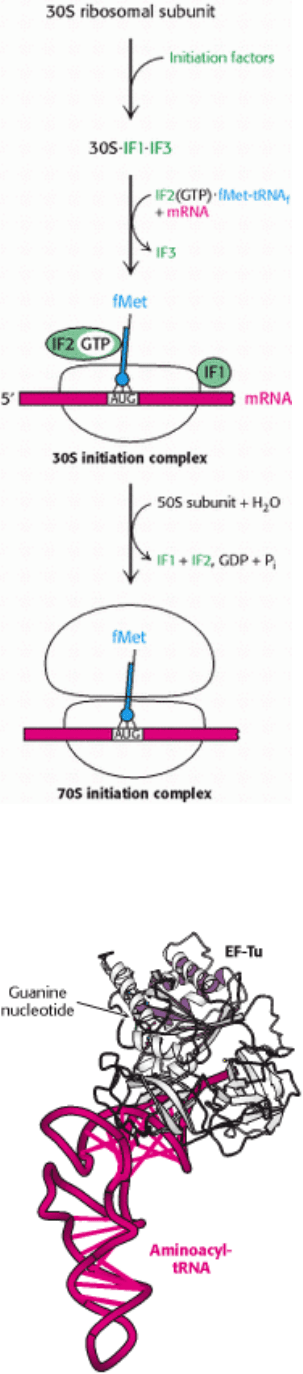
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.4. Protein Factors Play Key Roles in Protein Synthesis
Figure 29.27. Translation Initiation in Prokaryotes. Initiation factors aid the assembly first of the 30S initiation
complex and then of the 70S initiation complex.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.4. Protein Factors Play Key Roles in Protein Synthesis
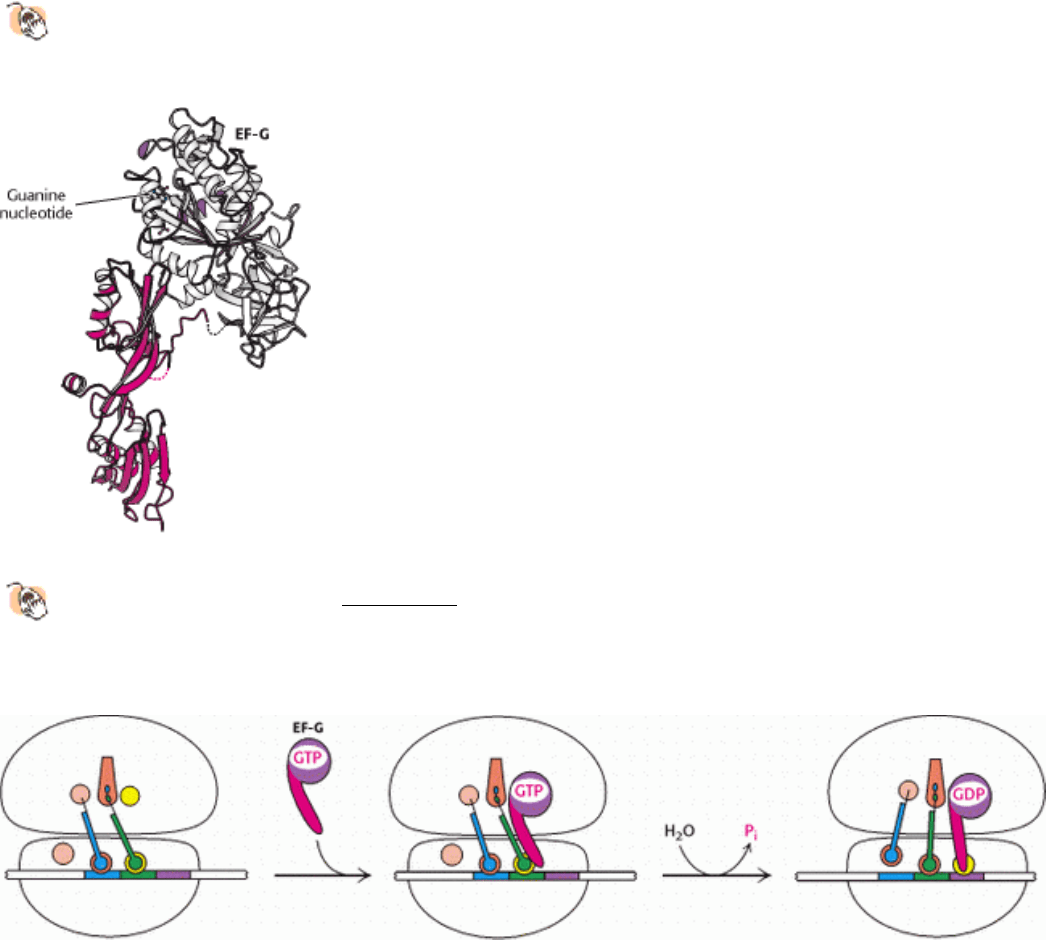
Figure 29.28. Structure of Elongation Factor Tu. The structure of a complex between elongation factor Tu (EF-Tu)
and an aminoacyl-tRNA. The amino-terminal domain of EF-Tu is a P-loop NTPase domain similar to those in
other G proteins.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.4. Protein Factors Play Key Roles in Protein Synthesis
Figure 29.29. Molecular Mimicry.
The structure of elongation factor G (EF-G) is remarkably similar in shape to that of
the EF-Tu-tRNA complex (see Figure 29.28). The amino-terminal region of EF-G is homologous to EF-Tu, and
the carboxyl-terminal region (shown in red) comprises a set of protein domains that adopted the shape of a tRNA
molecule over the course of evolution.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.4. Protein Factors Play Key Roles in Protein Synthesis
Figure 29.30. Translocation Mechanism. In the GTP form, EF-G binds to the EF-Tu-binding site on the 50S subunit.
This stimulates GTP hydrolysis, inducing a conformational change in EF-G, and driving the stem of EF-G into the A site
on the 30S subunit. To accommodate this domain, the tRNAs and mRNA move through the ribosome by a distance
corresponding to one codon.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.4. Protein Factors Play Key Roles in Protein Synthesis
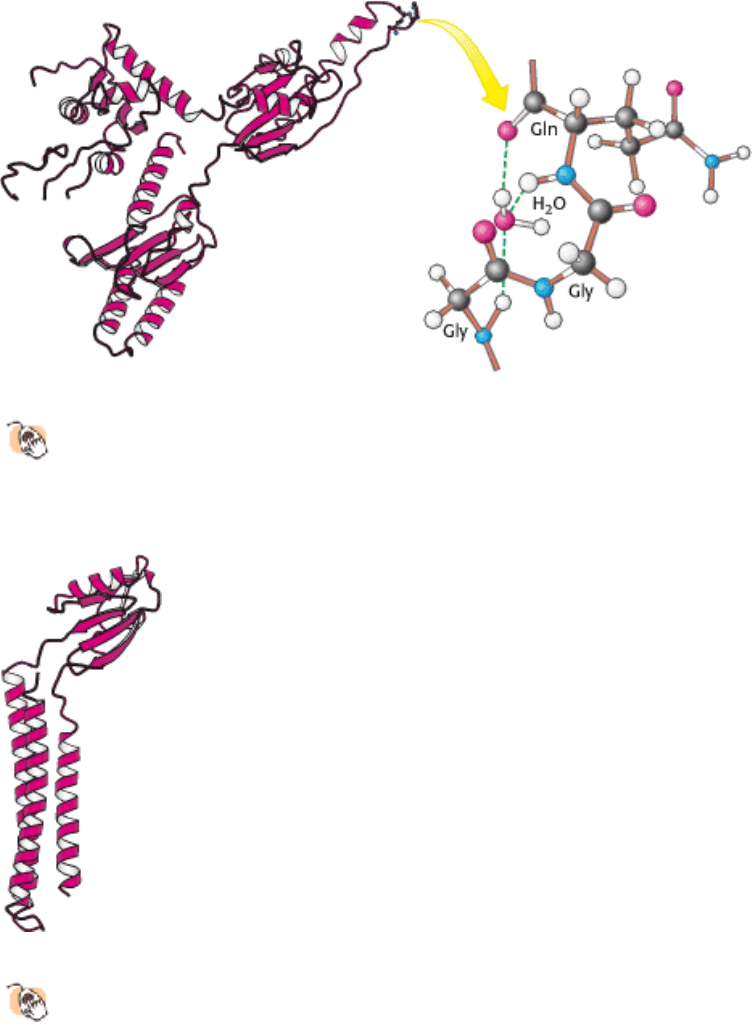
Figure 29.31. Structure of a Release Factor.
The structure of a eukaryotic release factor reveals a tRNA-like fold. The
acceptor-stem mimic includes the sequence Gly-Gly-Gln at its tip. This region appears to bind a water molecule,
which may be brought into the peptidyl transferase center. There it can participate in the cleavage of the peptidyl-
tRNA ester bond, with the aid of the glutamine residue and the ribosomal catalytic apparatus.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.4. Protein Factors Play Key Roles in Protein Synthesis
Figure 29.32. Structure of Ribosome Release Factor (RRF).
RRF is another protein that resembles tRNA. The α
helices of this protein mimic the tRNA structure. In contrast, in EF-G, β strands are the mimics, revealing an
independent evolutionary origin.
III. Synthesizing the Molecules of Life 29. Protein Synthesis
29.5. Eukaryotic Protein Synthesis Differs from Prokaryotic Protein Synthesis
Primarily in Translation Initiation
The basic plan of protein synthesis in eukaryotes and archaea is similar to that in bacteria. The major structural and
mechanistic themes recur in all domains of life. However, eukaryotic protein synthesis entails more protein components
than does prokaryotic protein synthesis, and some steps are more intricate. Some noteworthy similarities and differences
are as follows:
1. Ribosomes. Eukaryotic ribosomes are larger. They consist of a 60S large subunit and a 40S small subunit, which come
together to form an 80S particle having a mass of 4200 kd, compared with 2700 kd for the prokaryotic 70S ribosome.
The 40S subunit contains an 18S RNA that is homologous to the prokaryotic 16S RNA. The 60S subunit contains three
RNAs: the 5S and 28S RNAs are the counterparts of the prokaryotic 5S and 23S molecules; its 5.8S RNA is unique to

eukaryotes.
2. Initiator tRNA. In eukaryotes, the initiating amino acid is methionine rather than N-formylmethionine. However, as in
prokaryotes, a special tRNA participates in initiation. This aminoacyl-tRNA is called Met-tRNA
i
or Met-tRNA
f
(the
subscript "i" stands for initiation, and "f" indicates that it can be formylated in vitro).
3. Initiation. The initiating codon in eukaryotes is always AUG. Eukaryotes, in contrast with prokaryotes, do not use a
specific purine-rich sequence on the 5 side to distinguish initiator AUGs from internal ones. Instead, the AUG nearest
the 5
end of mRNA is usually selected as the start site. A 40S ribosome attaches to the cap at the 5 end of eukaryotic
mRNA (Section 28.3.1) and searches for an AUG codon by moving step-by-step in the 3 direction (Figure 29.33). This
scanning process in eukaryotic protein synthesis is powered by helicases that hydrolyze ATP. Pairing of the anticodon of
Met-tRNA
i
with the AUG codon of mRNA signals that the target has been found. In almost all cases, eukaryotic mRNA
has only one start site and hence is the template for a single protein. In contrast, a prokaryotic mRNA can have multiple
Shine-Dalgarno sequences and, hence, start sites, and it can serve as a template for the synthesis of several proteins.
Eukaryotes utilize many more initiation factors than do prokaryotes, and their interplay is much more intricate. The
prefix eIF denotes a eukaryotic initiation factor. For example, eIF-4E is a protein that binds directly to the 7-
methylguanosine cap (Section 28.3.1), whereas eIF-4A is a helicase. The difference in initiation mechanism between
prokaryotes and eukaryotes is, in part, a consequence of the difference in RNA processing. The 5 end of mRNA is
readily available to ribosomes immediately after transcription in prokaryotes. In contrast, pre-mRNA must be processed
and transported to the cytoplasm in eukaryotes before translation is initiated. Thus, there is ample opportunity for the
formation of complex secondary structures that must be removed to expose signals in the mature mRNA. The 5
cap
provides an easily recognizable starting point. In addition, the complexity of eukaryotic translation initiation provides
another mechanism for gene expression that we shall explore further in Chapter 31.
4. Elongation and termination. Eukaryotic elongation factors EF1α and EF1β γ are the counterparts of prokaryotic EF-
Tu and EF-Ts. The GTP form of EF1α delivers aminoacyl-tRNA to the A site of the ribosome, and EF1β γ catalyzes the
exchange of GTP for bound GDP. Eukaryotic EF2 mediates GTP-driven translocation in much the same way as does
prokaryotic EF-G. Termination in eukaryotes is carried out by a single release factor, eRF1, compared with two in
prokaryotes. Finally, eIF3, like its prokaryotic counterpart IF3, prevents the reassociation of ribosomal subunits in the
absence of an initiation complex.
29.5.1. Many Antibiotics Work by Inhibiting Protein Synthesis
The differences between eukaryotic and prokaryotic ribosomes can be exploited for the development of antibiotics
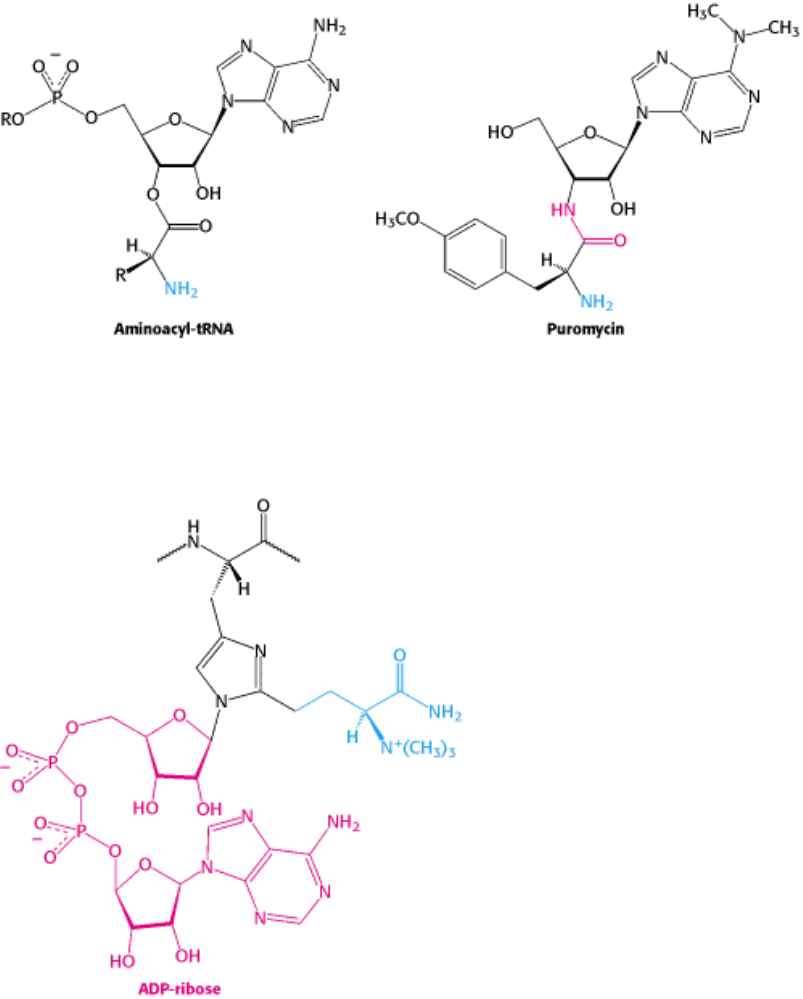
(Table 29.4). For example, the antibiotic puromycin inhibits protein synthesis by causing nascent prokaryotic
polypeptide chains to be released before their synthesis is completed. Puromycin is an analog of the terminal aminoacyl-
adenosine part of aminoacyl-tRNA (Figure 29.34).
It binds to the A site on the ribosome and inhibits the entry of aminoacyl-tRNA. Furthermore, puromycin contains an α-
amino group. This amino group, like the one on aminoacyl-tRNA, forms a peptide bond with the carboxyl group of the
growing peptide chain. The product, a peptide having a covalently attached puromycin residue at its carboxyl end,
dissociates from the ribosome.
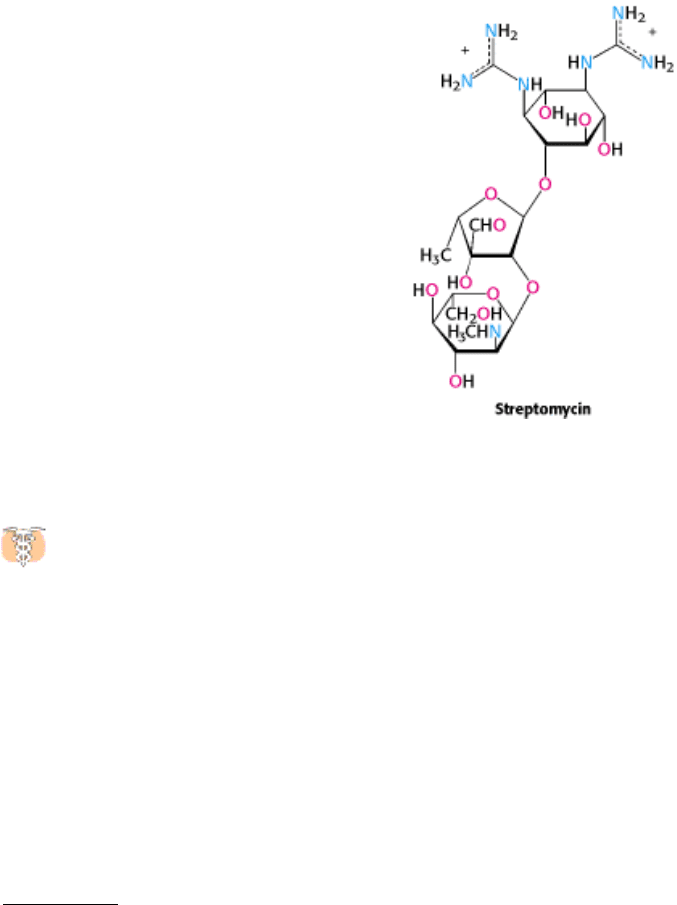
Streptomycin, a highly basic trisaccharide, interferes with the binding of formylmethionyl-tRNA to ribosomes and
thereby prevents the correct initiation of protein synthesis. Other aminoglycoside antibiotics such as neomycin,
kanamycin, and gentamycin interfere with the decoding site located near nucleotide 1492 in 16S rRNA of the 30S
subunit (Section 29.3.9). Chloramphenicol acts by inhibiting peptidyl transferase activity. Erythromycin binds to the 50S
subunit and blocks translocation. Finally, cyclohexamide blocks peptidyl transferase activity in eukaryotic ribosomes,
making a useful laboratory tool for blocking protein synthesis in eukaryotic cells.
29.5.2. Diphtheria Toxin Blocks Protein Synthesis in Eukaryotes by Inhibiting
Translocation
Diphtheria was a major cause of death in childhood before the advent of effective immunization. The lethal effects
of this disease are due mainly to a protein toxin produced by Corynebacterium diphtheriae, a bacterium that grows
in the upper respiratory tract of an infected person. The gene that encodes the toxin comes from a lysogenic phage that is
harbored by some strains of C. diphtheriae. A few micrograms of diphtheria toxin is usually lethal in an unimmunized
person because it inhibits protein synthesis. The toxin is cleaved shortly after entering a target cell into a 21-kd A
fragment and a 40-kd B fragment. The A fragment of the toxin catalyzes the covalent modification of an important
component of the protein-synthesizing machinery, whereas the B fragment enables the A fragment to enter the cytosol of
its target cell.
A single A fragment of the toxin in the cytosol can kill a cell. Why is it so lethal? The target of the A fragment is EF2,
the elongation factor catalyzing translocation in eukaryotic protein synthesis. EF2 contains diphthamide, an unusual
amino acid residue of unknown function that is formed by posttranslational modification of histidine. The A fragment
catalyzes the transfer of the adenosine diphosphate ribose unit of NAD
+
to a nitrogen atom of the diphthamide ring
(Figure 29.35). This ADP-ribosylation of a single side chain of EF2 blocks its capacity to carry out translocation of the
growing polypeptide chain. Protein synthesis ceases, accounting for the remarkable toxicity of diphtheria toxin.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.5. Eukaryotic Protein Synthesis Differs from Prokaryotic Protein Synthesis Primarily in Translation Initiation
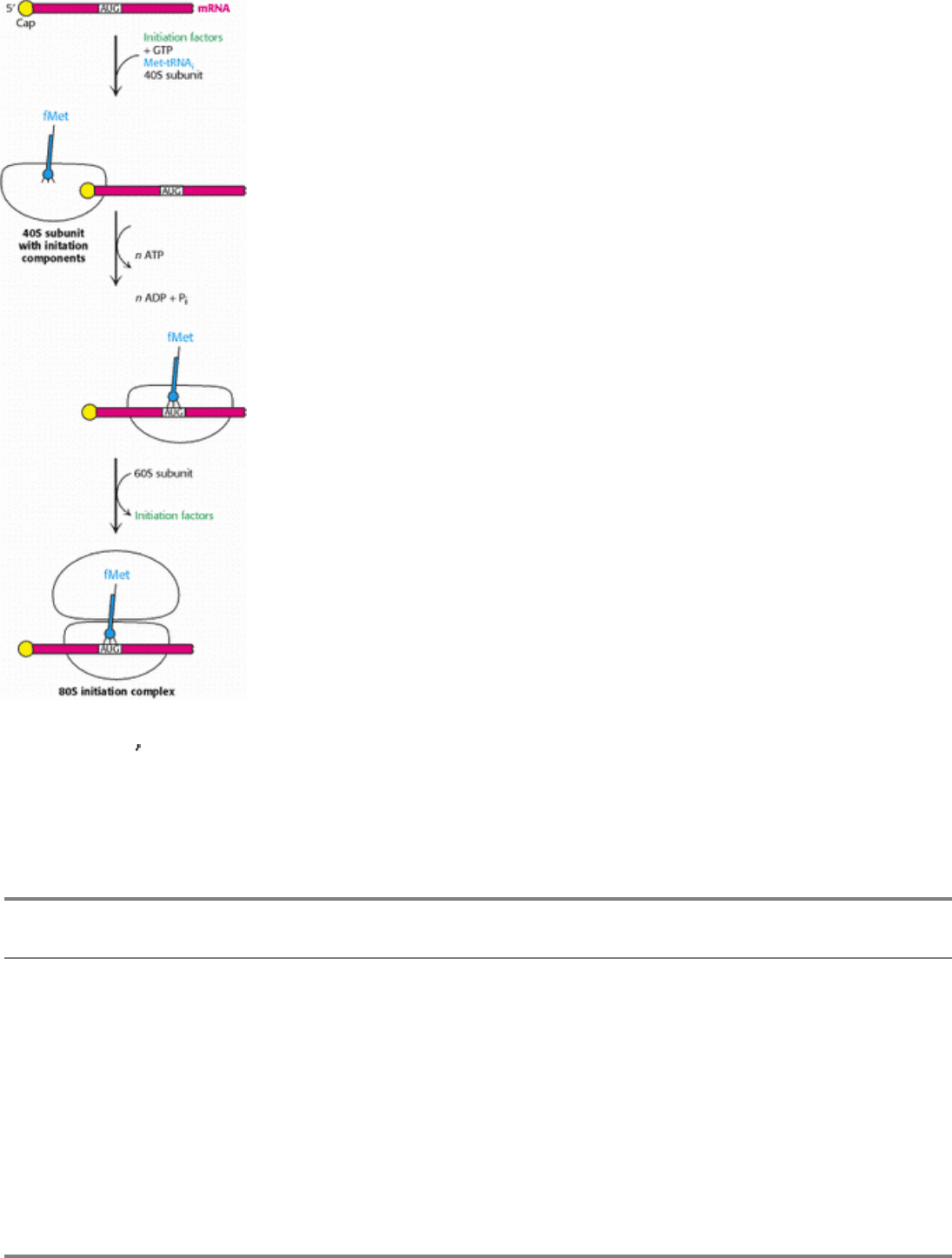
Figure 29.33. Eukaryotic Translation Initiation. In eukaryotes, translation initiation starts with the assembly of a
complex on the 5
cap that includes the 40S subunit and Met-tRNA
i
. Driven by ATP hydrolysis, this complex scans the
mRNA until the first AUG is reached. The 60S subunit is then added to form the 80S initiation complex.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.5. Eukaryotic Protein Synthesis Differs from Prokaryotic Protein Synthesis Primarily in Translation Initiation
Table 29.4. Antibiotic inhibitors of protein synthesis
Antibiotic Action
Streptomycin and other aminoglycosides Inhibit initiation and cause misreading of mRNA (prokaryotes)
Tetracycline Binds to the 30S subunit and inhibits binding of aminoacyl-tRNAs
(prokaryotes)
Chloramphenicol Inhibits the peptidyl transferase activity of the 50S ribosomal subunit
(prokaryotes)
Cycloheximide Inhibits the peptidyl transferase activity of the 60S ribosomal subunit
(eukaryotes)
Erythromycin Binds to the 50S subunit and inhibits translocation (prokaryotes)
Puromycin Causes premature chain termination by acting as an analog of aminoacyl-
tRNA (prokaryotes and eukaryotes)
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.5. Eukaryotic Protein Synthesis Differs from Prokaryotic Protein Synthesis Primarily in Translation Initiation
Figure 29.34. Antibiotic Action of Puromycin. Puromycin resembles the aminoacyl terminus of an aminoacyl-tRNA.
Its amino group joins the carbonyl group of the growing polypeptide chain to form an adduct that dissociates from the
ribosome. This adduct is stable because puromycin has an amide (shown in red) rather than an ester linkage.
III. Synthesizing the Molecules of Life 29. Protein Synthesis 29.5. Eukaryotic Protein Synthesis Differs from Prokaryotic Protein Synthesis Primarily in Translation Initiation
Figure 29.35. Blocking of Translocation by Diphtheria Toxin. Diphtheria toxin blocks protein synthesis in eukaryotes
by catalyzing the transfer of an ADP-ribose unit from NAD
+
to diphthamide, a modified amino acid residue in
elongation factor 2 (translocase). Diphthamide is formed by a posttranslational modification (blue) of a histidine residue.

III. Synthesizing the Molecules of Life 29. Protein Synthesis
Summary
Protein Synthesis Requires the Translation of Nucleotide Sequences into Amino Acid
Sequences
Protein synthesis is called translation because information present as a nucleic acid sequence is translated into a different
language, the sequence of amino acids in a protein. This complex process is mediated by the coordinated interplay of
more than a hundred macromolecules, including mRNA, rRNAs, tRNAs, aminoacyl-tRNA synthetases, and protein
factors. Given that proteins typically comprise from 100 to 1000 amino acids, the frequency at which an incorrect amino
acid is incorporated in the course of protein synthesis must be less than 10
-4
. Transfer RNAs are the adaptors that make
the link between a nucleic acid and an amino acid. These molecules, single chains of about 80 nucleotides, have an L-
shaped structure.
Aminoacyl-Transfer-RNA Synthetases Read the Genetic Code
Each amino acid is activated and linked to a specific transfer RNA by an enzyme called an aminoacyl-tRNA synthetase.
Such an enzyme links the carboxyl group of an amino acid to the 2
- or 3 -hydroxyl group of the adenosine unit of a CCA
sequence at the 3
end of the tRNA by an ester linkage. There is at least one specific aminoacyl-tRNA synthetase and at
least one specific tRNA for each amino acid. A synthetase utilizes both the functional groups and the shape of its cognate
amino acid to prevent the attachment of an incorrect amino acid to a tRNA. Some synthetases have a separate active site
at which incorrectly linked amino acids are removed by hydrolysis. A synthetase recognizes the anticodon, the acceptor
stem, and sometimes other parts of its tRNA substrate. By specifically recognizing both amino acids and tRNAs,
aminoacyl-tRNA synthetases implement the instruction of the genetic code. There exist two evolutionary distinct classes
of synthetases, each recognizing 10 amino acids. The two classes recognize opposite faces of tRNA molecules.
A Ribosome Is a Ribonucleoprotein Particle (70S) Made of a Small (30S) and a Large
(50S) Subunit
Protein synthesis takes place on ribosomes
ribonucleoprotein particles (about two-thirds RNA and one-third protein)
consisting of large and small subunits. In E. coli, the 70S ribosome (2700 kd) is made up of 30S and 50S subunits. The
30S subunit consists of 16S ribosomal RNA and 21 different proteins; the 50S subunit consists of 23S and 5S rRNA and
34 different proteins. The structure of almost all components of the ribosome have now been determined at or near
atomic resolution.
Proteins are synthesized in the amino-to-carboxyl direction, and mRNA is translated in the 5
3 direction. The start
signal on prokaryotic mRNA is AUG (or GUG) preceded by a purine-rich sequence that can base-pair with 16S rRNA.
In prokaryotes, transcription and translation are closely coupled. Several ribosomes can simultaneously translate an
mRNA, forming a polysome.
The ribosome includes three sites for tRNA binding called the A (aminoacyl) site, the P (peptidyl) site, and the E (exit)
site. With a tRNA attached to the growing peptide chain in the P site, an aminoacyl-tRNA binds to the A site. A peptide
bond is formed when the amino group of the aminoacyl-tRNA nucleophically attacks the ester carbonyl group of the
peptidyl-tRNA. On peptide-bond formation, the tRNAs and mRNA must be translocated for the next cycle to begin. The
deacylated tRNA moves to the E site and then leaves the ribosome, and the peptidyl-tRNA moves from the A site into
the P site.
The codons of messenger RNA recognize the anticodons of trans-fer RNAs rather than the amino acids attached to the
tRNAs. A codon on mRNA forms base pairs with the anticodon of the tRNA. Some tRNAs are recognized by more than
one codon because pairing of the third base of a codon is less crucial than that of the other two (the wobble mechanism).
