Berg J.M., Tymoczko J.L., Stryer L. Biochemistry
Подождите немного. Документ загружается.


Defects in other repair systems can increase the frequency of other tumors. For example, hereditary nonpolyposis
colorectal cancer (HNPCC, or Lynch syndrome) results from defective DNA mismatch repair. HNPCC is not rare as
many as 1 in 200 people will develop this form of cancer. Mutations in two genes, called hMSH2 and hMLH1, account
for most cases of this hereditary predisposition to cancer. The striking finding is that these genes encode the human
counterparts of MutS and MutL of E. coli. The MutS protein binds to mismatched base pairs (e.g., G-T) in DNA. An
MutH protein, together with MutL, participates in cleaving one of the DNA strands in the vicinity of this mismatch to
initiate the repair process (Figure 27.51). It seems likely that mutations in hMSH2 and hMLH1 lead to the accumulation
of mutations throughout the genome. In time, genes important in controlling cell proliferation become altered, resulting
in the onset of cancer.
27.6.6. Some Genetic Diseases Are Caused by the Expansion of Repeats of Three
Nucleotides
Some genetic diseases are caused by the presence of DNA sequences that are inherently prone to errors in the
course of replication. A particularly important class of such diseases are characterized by the presence of long
tandem arrays of repeats of three nucleotides. An example is Hunt-ington disease, an autosomal dominant neurological
disorder with a variable age of onset. The mutated gene in this disease expresses a protein called huntingtin, which is
expressed in the brain and contains a stretch of consecutive glutamine residues. These glutamine residues are encoded by
a tandem array of CAG sequences within the gene. In unaffected persons, this array is between 6 and 31 repeats,
whereas, in those with the disease, the array is between 36 and 82 repeats or longer. Moreover, the array tends to become
longer from one generation to the next. The consequence is a phenomenon called anticipation: the children of an
affected parent tend to show symptoms of the disease at an earlier age than did the parent.
The tendency of these trinucleotide repeats to expand is explained by the formation of alternative structures in DNA
replication (Figure 27.52). Part of the array within the daughter strand can loop out without disrupting base-pairing
outside this region. DNA polymerase extends this strand through the remainder of the array, leading to an increase in the
number of copies of the trinucleotide sequence.
A number of other neurological diseases are characterized by expanding arrays of trinucleotide repeats. How do these
long stretches of repeated amino acids cause disease? For huntingtin, it appears that the polyglutamine stretches become
increasingly prone to aggregate as their length increases; the additional consequences of such aggregation are still under
active investigation.
27.6.7. Many Potential Carcinogens Can Be Detected by Their Mutagenic Action on
Bacteria
Many human cancers are caused by exposure to chemicals. These chemical carcinogens usually cause mutations, which
suggests that damage to DNA is a fundamental event in the origin of mutations and cancer. It is important to identify
these compounds and ascertain their potency so that human exposure to them can be minimized. Bruce Ames devised a
simple and sensitive test for detecting chemical mutagens. In the Ames test, a thin layer of agar containing about 10
9
bacteria of a specially constructed tester strain of Salmonella is placed on a petri dish. These bacteria are unable to grow
in the absence of histidine, because a mutation is present in one of the genes for the biosynthesis of this amino acid. The
addition of a chemical mutagen to the center of the plate results in many new mutations. A small proportion of them
reverse the original mutation, and histidine can be synthesized. These revertants multiply in the absence of an external
source of histidine and appear as discrete colonies after the plate has been incubated at 37°C for 2 days (Figure 27.53).
For example, 0.5 µg of 2-aminoanthracene gives 11,000 revertant colonies, compared with only 30 spontaneous
revertants in its absence. A series of concentrations of a chemical can be readily tested to generate a dose-response curve.
These curves are usually linear, which suggests that there is no threshold concentration for mutagenesis.
Some of the tester strains are responsive to base-pair substitutions, whereas others detect deletions or additions of base
pairs (frameshifts). The sensitivity of these specially designed strains has been enhanced by the genetic deletion of their
excision-repair systems. Potential mutagens enter the tester strains easily because the lipopolysaccharide barrier that
normally coats the surface of Salmonella is incomplete in these strains.
A key feature of this detection system is the inclusion of a mammalian liver homogenate (Section 4.1.2). Recall that
some potential carcinogens such as aflatoxin are converted into their active forms by enzyme systems in the liver or
other mammalian tissues (Section 27.6.1). Bacteria lack these enzymes, and so the test plate requires a few milligrams of
a liver homogenate to activate this group of mutagens.
The Salmonella test is extensively used to help evaluate the mutagenic and carcinogenic risks of a large number of
chemicals. This rapid and inexpensive bacterial assay for mutagenicity complements epidemiological surveys and animal
tests that are necessarily slower, more laborious, and far more expensive. The Salmonella test for mutagenicity is an
outgrowth of studies of gene-protein relations in bacteria. It is a striking example of how fundamental research in
molecular biology can lead directly to important advances in public health.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
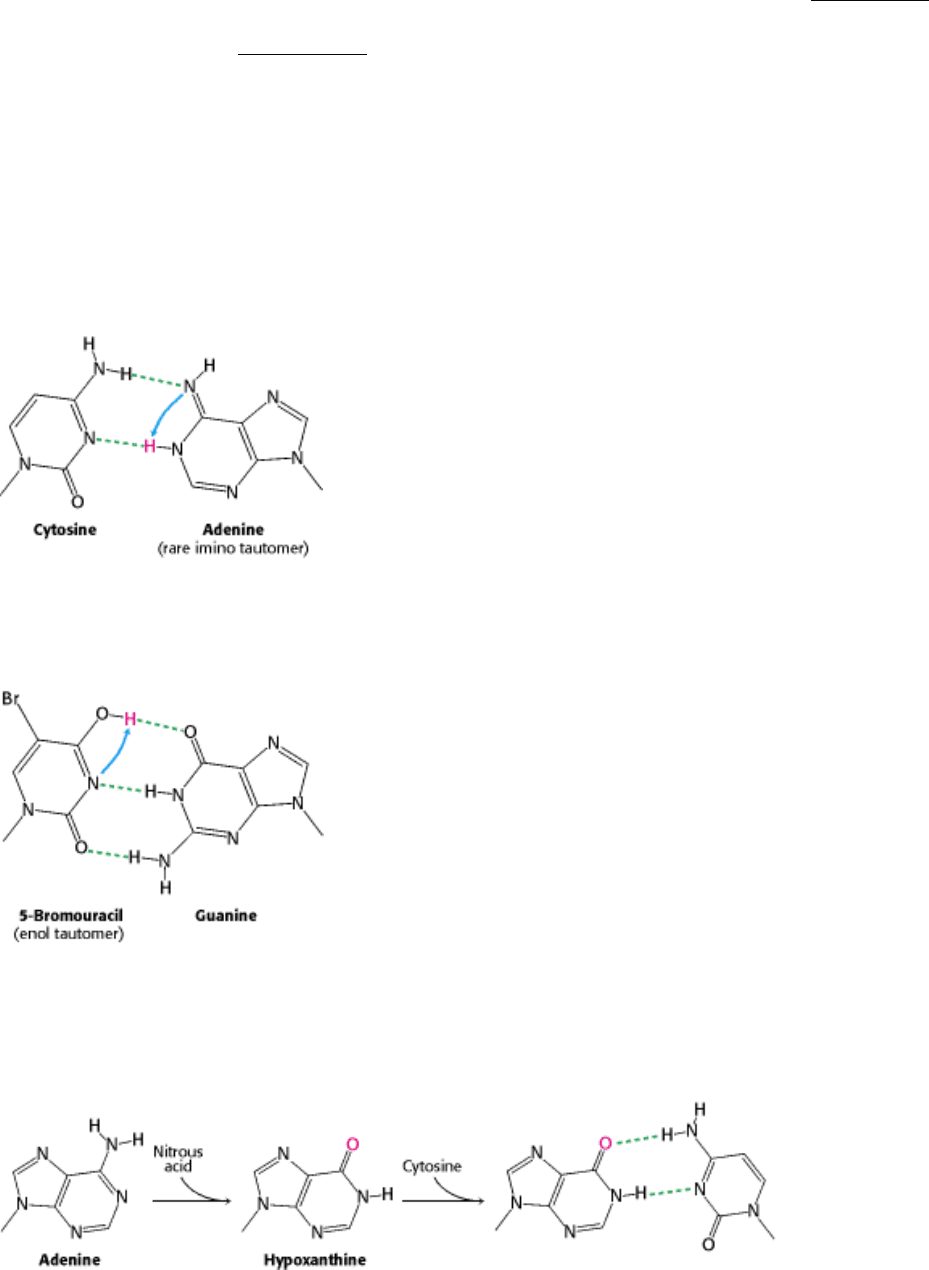
Figure 27.41. Base Pair with Mutagenic Tautomer. The bases of DNA can exist in rare tautomeric forms. The imino
tautomer of adenine can pair with cytosine, eventually leading to a transition from A-T to G-C.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
Figure 27.42. Base Pair with 5-Bromouracil. This analog of thymine has a higher tendency to form an enol tautomer
than does thymine itself. The pairing of the enol tautomer of 5-bromouracil with guanine will lead to a transition from T-
A to C-G.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
Figure 27.43. Chemical Mutagenesis. Treatment of DNA with nitrous acid results in the conversion of adenine into
hypoxanthine. Hypoxanthine pairs with cytosine, inducing a transition from A-T to G-C.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
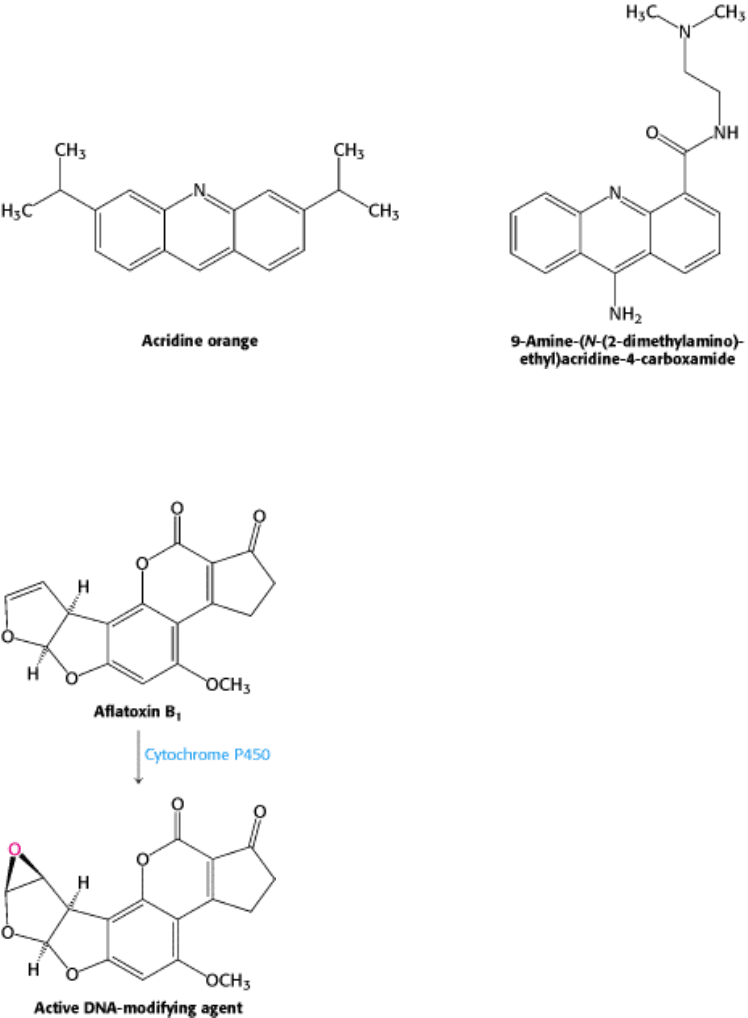
Figure 27.44. Acridines. Acridine dyes induce frameshift mutations by intercalating into the DNA, leading to the
incorporation of an additional base on the opposite strand.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
Figure 27.45. Aflatoxin Reaction. The compound, produced by molds that grow on peanuts, is activated by cytochrome
P450 to form a highly reactive species that modifies bases such as guanine in DNA, leading to mutations.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
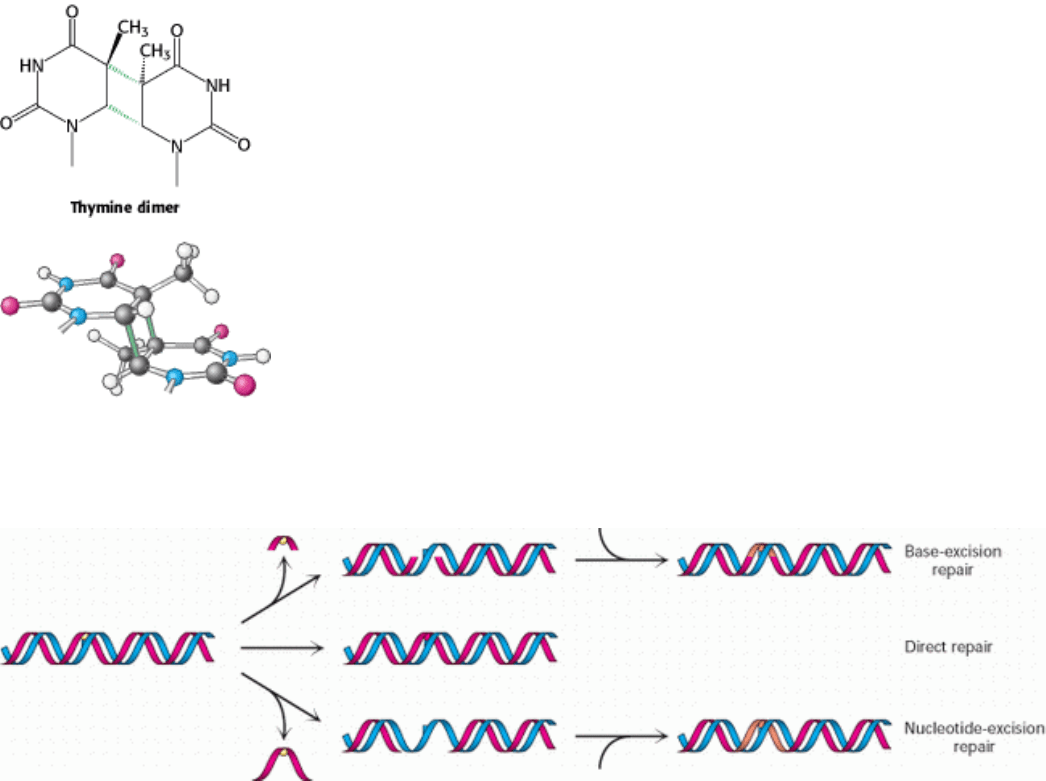
Figure 27.46. Cross-Linked Dimer of Two Thymine Bases. Ultraviolet light induces cross-links between adjacent
pyrimidines along one strand of DNA.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
Figure 27.47. Repair Pathways. Three different pathways are used to repair damaged regions in DNA. In base-excision
repair, the damaged base is removed and replaced. In direct repair, the damaged region is corrected in place. In
nucleotide-excision repair, a stretch of DNA around the site of damage is removed and replaced.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
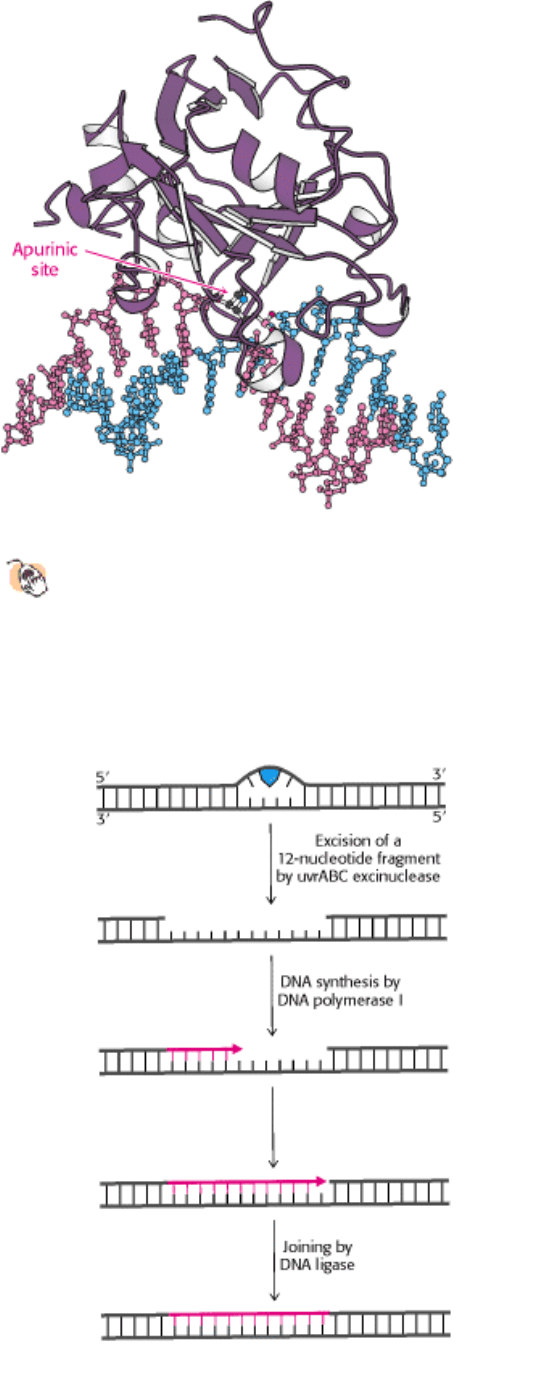
Figure 27.48. Structure of DNA-Repair Enzyme.
A complex between the DNA-repair enzyme AlkA and an analog of
an apurinic site. Note that the damaged base is flipped out of the DNA double helix into the active site of the
enzyme for excision.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
Figure 27.49. Excision Repair. Repair of a region of DNA containing a thymine dimer by the sequential action of a
specific excinuclease, a DNA polymerase, and a DNA ligase. The thymine dimer is shown in blue, and the new region of
DNA is in red. [After P. C. Hanawalt. Endevour 31(1982):83.]
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
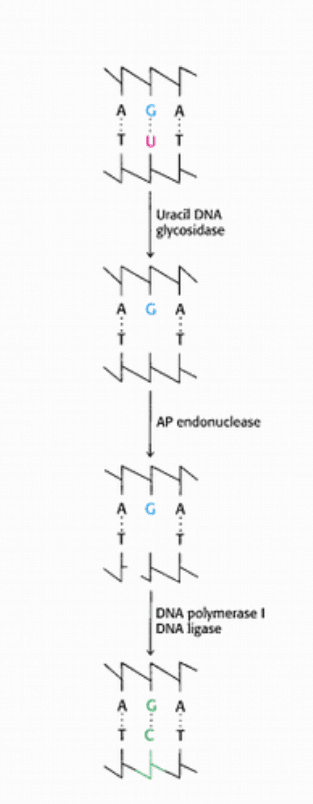
Figure 27.50. Uracil Repair. Uracil bases in DNA, formed by the deamination of cytosine, are excised and replaced by
cytosine.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
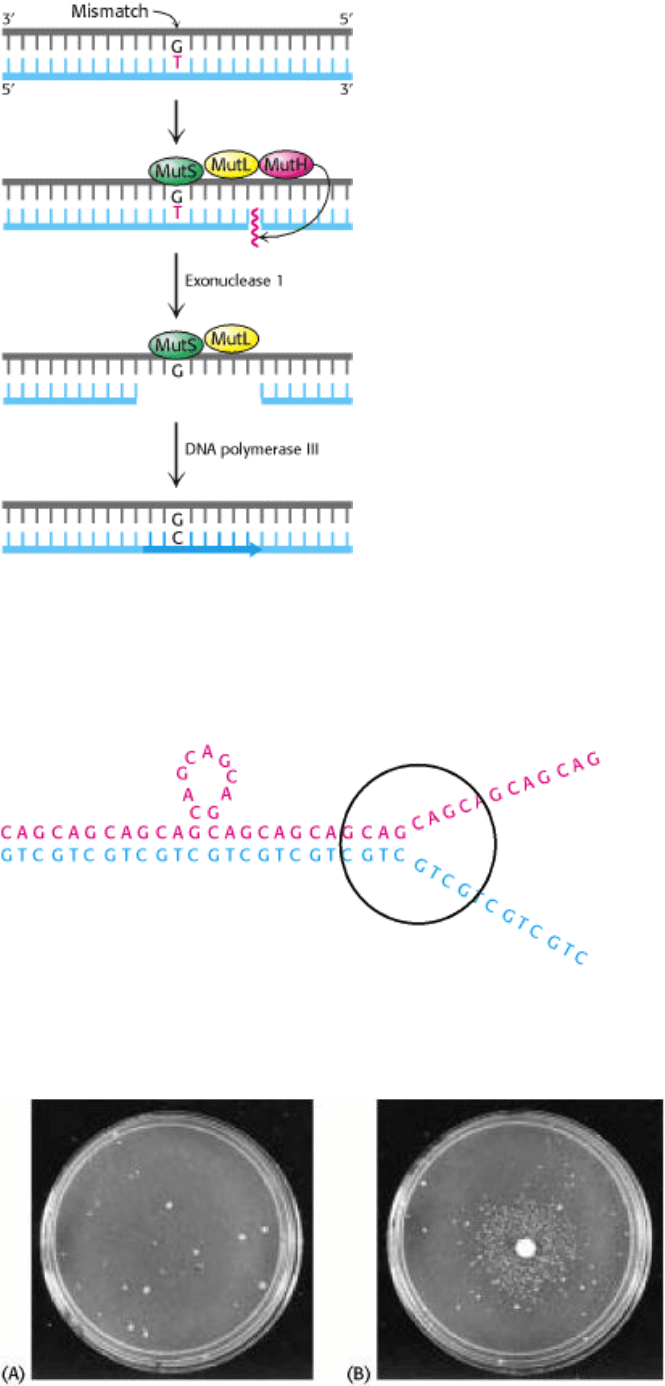
Figure 27.51. Mismatch Repair. DNA mismatch repair in E. coli is initiated by the interplay of MutS, MutL, and MutH
proteins. A G-T mismatch is recognized by MutS. MutH cleaves the backbone in the vicinity of the mismatch. A
segment of the DNA strand containing the erroneous T is removed by exonuclease I and synthesized anew by DNA
polymerase III. [After R. F. Service. Science 263(1994):1559.]
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
Figure 27.52. Triplet Repeat Expansion. Sequences containing tandem arrays of repeated triplet sequences can be
expanded to include more repeats by the looping out of some of the repeats before replication.
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair 27.6. Mutations Involve Changes in the Base Sequence of DNA
Figure 27.53. Ames Test. (A) A petri dish containing about 10
9
Salmonella bacteria that cannot synthesize histidine and
(B) a petri dish containing a filter-paper disc with a mutagen, which produces a large number of revertants that can
synthesize histidine. After 2 days, the revertants appear as rings of colonies around the disc. The small number of visible
colonies in plate A are spontaneous revertants. [From B. N. Ames, J. McCann, and E. Yamasaki. Mutat. Res. 31
(1975):347.]
III. Synthesizing the Molecules of Life 27. DNA Replication, Recombination, and Repair
Summary
DNA Can Assume a Variety of Structural Forms
DNA is a structurally dynamic molecule that can exist in a variety of helical forms: A-DNA, B-DNA (the classic Watson-
Crick helix), and Z-DNA. DNA can be bent, kinked, and unwound. In A-, B-, and Z-DNA, two antiparallel chains are
held together by Watson-Crick base pairs and stacking interactions between bases in the same strand. The sugar-
phosphate backbone is on the outside, and the bases are inside the double helix. A- and B-DNA are right-handed helices.
In B-DNA, the base pairs are nearly perpendicular to the helix axis. In A-DNA, the bases are tilted rather than
perpendicular. An important structural feature of the B helix is the presence of major and minor grooves, which display
different potential hydrogen-bond acceptors and donors according to the base sequence. X-ray analysis of a single crystal
of B-DNA reveals that the structure is much more variable than was originally imagined. Dehydration induces the
transition from B- to A-DNA. Z-DNA is a left-handed helix. It can be formed in regions of DNA in which purines
alternate with pyrimidines, as in CGCG or CACA. Most of the DNA in a cell is in the B-form.
DNA Polymerases Require a Template and a Primer
DNA polymerases are template-directed enzymes that catalyze the formation of phosphodiester bonds by the 3
-hydroxyl
group's nucleophilic attack on the innermost phosphorus atom of a deoxyribonucleoside 5
-triphosphate. They cannot
start chains de novo; a primer with a free 3
-hydroxyl group is required. DNA polymerases from a variety of sources
have important structural features in common as well as a catalytic mechanism requiring the presence of two metal ions.
Many DNA polymerases proofread the nascent product; their 3
5 exonuclease activity potentially edits the outcome
of each polymerization step. A mispaired nucleotide is excised before the next step proceeds. In E. coli, DNA
polymerase I repairs DNA and participates in replication. Fidelity is further enhanced by an induced fit that results in a
catalytically active conformation only when the complex of enzyme, DNA, and correct dNTP is formed. Helicases
prepare the way for DNA replication by using ATP hydrolysis to separate the strands of the double helix.
Double-Stranded DNA Can Wrap Around Itself to Form Supercoiled Structures
A key topological property of DNA is its linking number (Lk), which is defined as the number of times one strand of
DNA winds around the other in the right-hand direction when the DNA axis is constrained to lie in a plane. Molecules
differing in linking number are topoisomers of one another and can be interconverted only by cutting one or both DNA
strands; these reactions are catalyzed by topoisomerases. Changes in linking number generally lead to changes in both
the number of turns of double helix and the number of turns of superhelix. Topoisomerase II (DNA gyrase) catalyzes the
ATP-driven introduction of negative supercoils, which leads to the compaction of DNA and renders it more susceptible
to unwinding. Supercoiled DNA is relaxed by topoisomerase I. The unwinding of DNA at the replication fork is
catalyzed by an ATP-driven helicase.
DNA Replication of Both Strands Proceeds Rapidly from Specific Start Sites
DNA replication in E. coli starts at a unique origin (oriC) and proceeds sequentially in opposite directions. More than 20
proteins are required for replication. An ATP-driven helicase unwinds the oriC region to create a replication fork. At this
fork, both strands of parental DNA serve as templates for the synthesis of new DNA. A short stretch of RNA formed by
primase, an RNA polymerase, primes DNA synthesis. One strand of DNA (the leading strand) is synthesized
continuously, whereas the other strand (the lagging strand) is synthesized discontinuously, in the form of 1-kb fragments
(Okazaki fragments). Both new strands are formed simultaneously by the concerted actions of the highly processive

DNA polymerase III holoenzyme, an asymmetric dimer. The discontinuous assembly of the lagging strand enables 5
3
polymerization at the atomic level to give rise to overall growth of this strand in the 3 5 direction. The RNA
primer stretch is hydrolyzed by the 5
3 nuclease activity of DNA polymerase I, which also fills gaps. Finally,
nascent DNA fragments are joined by DNA ligase in a reaction driven by ATP or NAD
+
cleavage.
DNA synthesis in eukaryotes is more complex than in prokaryotes. Eukaryotes require thousands of origins of
replication to complete replication in a timely fashion. A special RNA-dependent DNA polymerase called telomerase is
responsible for the replication of the ends of linear chromosomes.
Double-Stranded DNA Molecules with Similar Sequences Sometimes Recombine
DNA molecules that are similar in nucleotide sequences in a local region can recombine to form DNA duplexes that
begin with the sequence of one molecule and continue with the sequence of the other. Recombinases catalyze the
formation of these products through the formation and resolution of Holliday junctions. In these structures, four DNA
strands come together to form a crosslike structure.
Recombinases cleave DNA strands and form specific adducts in which a tyrosine residue of the enzyme is linked to a 3
-
phosphoryl group of the DNA. These intermediates then react with 5
-hydroxyl groups of other DNA strands to form the
new phosphodiester bonds that are present in the recombination products. The reaction mechanisms of recombinases are
similar to those of type I topoisomerases.
Mutations Are Produced by Several Types of Changes in the Base Sequence of DNA
Mutations are produced by mistakes in base-pairing, by the covalent modification of bases, and by the deletion and
insertion of bases. The 3
5 exonuclease activity of DNA polymerases is critical in lowering the spontaneous
mutation rate, which arises in part from mispairing by tautomeric bases. Lesions in DNA are continually being repaired.
Multiple repair processes utilize information present in the intact strand to correct the damaged strand. For example,
pyrimidine dimers formed by the action of ultraviolet light are excised by uvrABC excinuclease, an enzyme that
removes a 12-nucleotide region containing the dimer. Xeroderma pigmentosum, a genetically transmitted disease, is
caused by the defective repair of lesions in DNA, such as pyrimidine dimers; patients with this disease usually develop
skin cancers. Many cancers of the colon are caused by defective DNA mismatch repair arising from mutations in human
genes that have been highly conserved in evolution. Damage to DNA is a fundamental event in both carcinogenesis and
mutagenesis. Many potential carcinogens can be detected by their mutagenic action on bacteria.
Key Terms
B-DNA helix
A-DNA helix
major groove
minor groove
Z-DNA helix
DNA polymerase
template
primer
exonuclease
helicase
supercoil
linking number (Lk)
topoisomer
twist (Tw)
writhe (Wr)
topoisomerase
origin of replication
primase
replication fork
Okazaki fragment
lagging strand
leading stand
DNA ligase
processivity
cell cycle
telomere
telomerase
recombinase
Holliday junction
recombination synapse
transition
transversion
mutagen
