Townsend C.R., Begon M., Harper J.L. Essentials of Ecology
Подождите немного. Документ загружается.

Chapter 7 Predation, grazing and disease
249
Predation, true predators, grazers and parasites
A predator may be defined as any organism that con-
sumes all or part of another living organism (its ‘prey’
or ‘host’) thereby benefiting itself, but, under at least
some circumstances, reducing the growth, fecundity
or survival of the prey.
‘True’ predators invariably kill their prey and do so
more or less immediately after attacking them, and
consume several or many prey items in the course
of their life. Grazers also attack several or many prey
items in the course of their life, but consume only part
of each prey item and do not usually kill their prey.
Parasites also consume only part of each host, and
also do not usually kill their host, especially in the short
term, but attack one or very few hosts in the course
of their life, with which they therefore often form a
relatively intimate association.
The subtleties of predation
Grazers and parasites, in particular, often exert their
harm not by killing their prey immediately like true
predators, but by making the prey more vulnerable
to some other form of mortality.
The effects of grazers and parasites on the organ-
isms they attack are often less profound than they first
seem because individual plants can compensate for
the effects of herbivory and hosts may have defensive
responses to attack by parasites.
The effects of predation on a population of prey
are complex to predict because the surviving prey
may experience reduced competition for a limiting
resource, or produce more offspring, or other pre-
dators may take fewer of the prey.
Predator behavior
True predators and grazers typically ‘forage’, moving
around within their habitat in search of their prey. Other
predators ‘sit and wait’ for their prey, though almost
always in a selected location. With parasites and
pathogens there may be direct transmission between
infectious and uninfected hosts, or contact between
free-living stages of the parasite and uninfected hosts
may be important.
Optimal foraging theory aims to understand why
particular patterns of foraging behavior have been
favored by natural selection (because they give rise
to the highest net rate of energy intake).
Generalist predators spend relatively little time
searching but include relatively low-profitability items
in their diet. Specialists only include high-profitability
items in their diet but spend a relatively large amount
of their time searching for them.
Population dynamics of predation
There is an underlying tendency for predators and
prey to exhibit cycles in abundance, and cycles are
observed in some predator–prey and host–parasite
interactions. However, there are many important
factors that can modify or override the tendency to
cycle.
Crowding of either predator or prey is likely to have
a damping effect on any predator–prey cycles.
Many populations of predators and prey exist as a
‘metapopulation’. In theory, and in practice, asynchrony
in population dynamics in different patches and the
process of dispersal tend to dampen any underlying
population cycles.
Predation and community structure
There are many situations where predation may hold
down the densities of populations, so that resources
are not limiting and individuals do not compete for
them. When predation promotes the coexistence of
species amongst which there would otherwise be
competitive exclusion (because the densities of some
or all of the species are reduced to levels at which
competition is relatively unimportant) this is known as
‘predator-mediated coexistence’.
The effects of predation generally on a group of
competing species depend on which species suffers
most. If it is a subordinate species, then this may be
driven to extinction and the total number of species
in the community will decline. If it is the competitive
dominants that suffer most, however, the results of
heavy predation will usually be to free space and
resources for other species, and species numbers
may then increase.
It is not unusual for the number of species in a
community to be greatest at intermediate levels of
predation.
SUMMARY
Summary
9781405156585_4_007.qxd 11/5/07 14:53 Page 249

Part III Individuals, Populations, Communities and Ecosystems
250
REVIEW QUESTIONS
Review questions
Asterisks indicate challenge questions
1 With the aid of examples, explain the feeding
characteristics of true predators, grazers,
parasites and parasitoids.
2* True predators, grazers and parasites can alter
the outcome of competitive interactions that
involve their ‘prey’ populations: discuss this
assertion using one example from each
category.
3 Discuss the various ways that plants may
‘compensate’ for the effects of herbivory.
4 Predation is ‘bad’ for the prey that get eaten.
Explain why it may be good for those that do
not get eaten.
5* Discuss the pros and cons, in energetic
terms, of (i) being a generalist as opposed
to a specialist predator, and (ii) being a
sit-and-wait predator as opposed to an
active forager.
6 In simple terms, explain why there is an
underlying tendency for populations of
predators and prey to cycle.
7* You have data that shows cycles in nature
among interacting populations of a true
predator, a grazer and a plant. Describe an
experimental protocol to determine whether this
is a grazer–plant cycle or a predator–grazer
cycle.
8 Define mutual interference and give examples
for true predators and parasites. Explain how
mutual interference may dampen inherent
population cycles.
9 Discuss the evidence presented in this chapter
that suggests environmental patchiness has
an important influence on predator–prey
population dynamics.
10 With the help of an example, explain why most
prey species may be found in communities
subject to an intermediate intensity of predation.
9781405156585_4_007.qxd 11/5/07 14:53 Page 250

251
Chapter 8
Evolutionary ecology
CHAPTER CONTENTS
8.1 Introduction
8.2 Molecular ecology: differentiation within and between species
8.3 Coevolutionary arms races
8.4 Mutualistic interactions
Chapter contents
KEY CONCEPTS
In this chapter you will:
l
appreciate the range of molecular (DNA) markers that have been used
in ecology
l
understand how these markers can be put to work in determining the
extent of subdivision within, and the degree of separation between,
species
l
recognize the importance of coevolutionary arms races in the
dynamics of the component populations, especially of plants
and their insect herbivores, and of parasites and their hosts
l
understand the nature of mutualistic interactions in general and their
crucial importance both for the species concerned and for almost all
communities on the planet
l
appreciate the particular contributions of mutualisms in diverse
areas from farming, through the functioning of guts and roots, to
the fixation of nitrogen by plants
Key concepts
9781405156585_4_008.qxd 11/5/07 14:54 Page 251

8.1 Introduction
In Chapter 2, we set the scene for the remainder of this book by illustrating
how, to modify slightly Dobzhansky’s famous phrase, ‘nothing in ecology makes
sense, except in the light of evolution’. But evolution does more than underpin
ecology (and the whole of the rest of biology). There are many areas in ecology
where evolutionary adaptation by natural selection takes center stage to the extent
that the term ‘evolutionary ecology’ is often used to describe them. In several
previous chapters, therefore, topics in evolutionary ecology have been dealt with,
quite naturally, as integral parts of broader ecological questions. In Chapter 3,
we examined the nature and importance of defenses that have evolved to protect
plants and prey from their predators. In Chapter 5, we saw how patterns in life
histories – schedules of growth, reproduction and so on – can only be understood
in relation to corresponding patterns in the habitats in which they have evolved.
In Chapter 6, we looked at interspecific competition as an evolutionary driving
force, generating patterns in the coexistence and exclusion of competing species.
And in Chapter 7, we discussed ‘optimal foraging’: the evolution of behavioral
strategies that maximize predator fitness and thus mold their dynamic interactions
with their prey.
This, of course, is not an exhaustive survey of topics in evolutionary ecology.
In the present chapter, therefore, we deal with a number of others (though the
final list will remain less than exhaustive). We focus especially on coevolution:
pairs of species acting as reciprocal driving forces in one another’s evolution. The
question of coevolutionary ‘arms races’ between predators and their prey is taken
up in Section 8.3, with a particular emphasis on host–pathogen interactions: each
adaptation in the prey that fends off or avoids the attacks of a predator provoking
a corresponding adaptation in the predator that improves its ability to overcome
those defenses. However, not all coevolutionary interactions are antagonistic.
Many species-pairs are ‘mutualists’: both parties benefiting, on balance at least,
from the interactions in which they take part. Some of the most important of
these mutualisms – pollination, corals and nitrogen fixation, for example – are
discussed in Section 8.4. We begin, though, not with species interactions but with
aspects of evolutionary differentiation within and between species, especially
those detectable by modern techniques developed in molecular genetics and thus
often described as aspects of ‘molecular ecology’.
Part III Individuals, Populations, Communities and Ecosystems
252
We have noted previously that nothing in ecology makes sense, except in the light
of evolution. But some areas of ecology are even more evolutionary than others.
We may need to look within individuals to examine the details of the genes
they carry, or to acknowledge explicitly the crucial and reciprocal role that
species play in one another’s evolution.
9781405156585_4_008.qxd 11/5/07 14:54 Page 252
8.2 Molecular ecology: differentiation within
and between species
For much of the time, it is entirely appropriate for ecologists to talk about
‘populations’ or ‘species’ as if they were singular, homogeneous entities: for
example, we may talk of ‘the distribution of Asian elephants’, saying nothing
about whether the species might be differentiated into distinct races or subgroups,
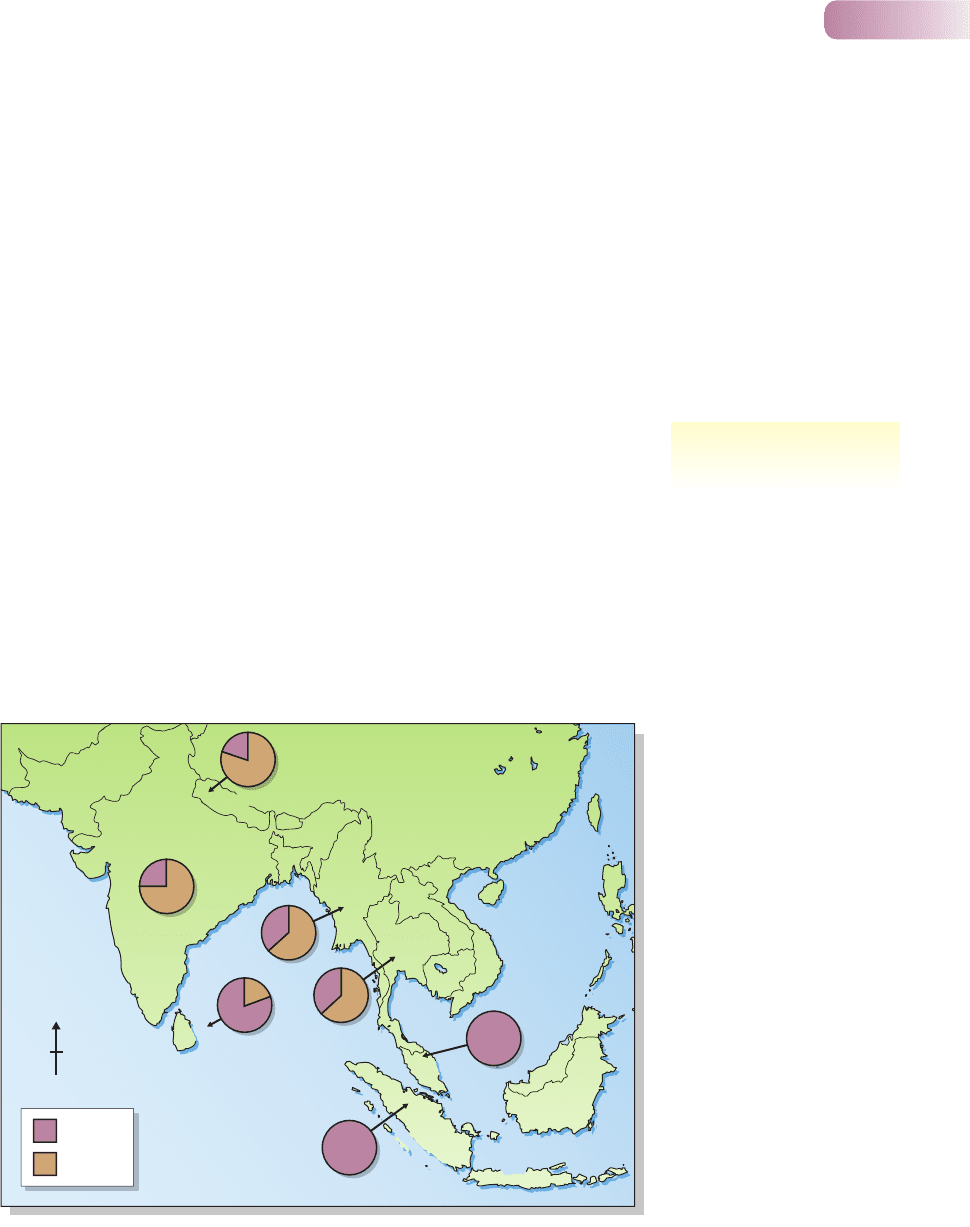
as indeed it is (Figure 8.1). But for some purposes, knowing how much differ-
entiation there is within species, or between one species and another, is critical
for an understanding of their dynamics, and ultimately for managing those
dynamics. Is a particular population derived largely from offspring born locally,
or from immigrants from another, distinguishable population? Where exactly
does the distribution of a particular species end and that of another, closely related
species begin? In cases like these, being able to determine, at a variety of scales,
who is most closely related to whom (and who is quite distinct from whom) may
be essential.
Our ability to do this itself depends on the resolution with which we can
differentiate individuals from one another and even determine where they came
from or who their parents were. In the past, this was difficult and frequently
impossible: reliance on simple, visual markers meant that all individuals within
a species often looked the same, and even members of closely related species
could often only be distinguished by experienced taxonomists looking down a
microscope at, say, details of a male’s genitalia. Now, though, molecular, genetic
markers (albeit still requiring experts and expensive equipment) have massively
Chapter 8 Evolutionary ecology
253
N
India
Arabian
Sea
Indian
Ocean
Sri
Lanka
Nepal
Clade A
Clade B
China
Myanmar
Thailand
Malaysia
Indonesia
(Sumatra)
Java
Figure 8.1
Distribution of two distinct ‘clades’ of
the Asian elephant, Elephas maximus
(groups with distinct evolutionary
histories following their common
origin), revealed only by an analysis
of molecular markers. These ciades
coexist in many areas, though their
distinctiveness itself suggests a
degree of independence in their
dynamics even when they do coexist.
AFTER FLEISCHER ET AL., 2001
the need to know who is most
closely related to whom
9781405156585_4_008.qxd 11/5/07 14:54 Page 253
increased the resolution at which we can differentiate between populations and
even between individuals, and hence have vastly improved our ability to address
these types of questions. We begin, therefore, in Box 8.1, with a brief survey of
some of the most important of these molecular markers and their uses.
Part III Individuals, Populations, Communities and Ecosystems
254
8.1 QUANTITATIVE ASPECTS
8.1 Quantitative aspects
This is not the place for crash courses in either
molecular biology or the laboratory methods used
to extract, amplify, separate and analyze molecular
markers, but it will nonetheless be useful to have
some appreciation of their nature and key properties
– and to be introduced to some of the technical terms
and abbreviations that abound in this area. Most
recent studies in ecology have used DNA of one type
or other for molecular identification. We need, at
the very least, to be aware that a length of DNA is
characterized by the sequence of bases of which it
is composed, adenine (A), cytosine (C), guanine (G)
and thymine (T), and that in double-stranded DNA,
these link across to one another in complementary
base-pairs: A-T and G-C.
Choosing a molecular marker
The basis for all uses of molecular markers in eco-
logy is that individuals can be differentiated from
one another to greater or lesser extents as a result
of molecular variation amongst them. The ultimate
source of this variation is mutation in the sequence
of bases, which, of course, occurs independently of
its consequences for the organism concerned. What
happens to the mutation, and the mutated organism,
then depends essentially on the balance between
selection and ‘genetic drift’ (random, undirected
changes in gene frequency from generation to gener-
ation). If the mutation occurs in a region of DNA that
is important because, say, it codes for a crucial part of
an essential enzyme, then selection is likely to deter-
mine the outcome. An unfavorable mutation (the vast
majority in important regions of DNA) will quickly be
lost because the mutated organism is less fit than its
counterparts. Individuals will therefore differ relatively
little in such regions, and if they do, differentiation is
most likely to reflect ‘adaptive’ variation: different vari-
ants being favored in different individuals, perhaps
because of where they live.
But there are also regions of DNA that appear not
to code for important parts of enzymes or to per-
form any other function where the precise sequence
is crucial. Variation in these regions is therefore said
to be ‘neutral’, and mutations can accumulate there
over time. Imagine two offspring of a single mating.
They will be genetically very similar. But imagine now
that each, literally, goes its own way. As each gener-
ation passes and mutations accumulate, the lineages
derived from them will become increasingly divergent
in those regions of their genome where variation is
neutral, and lineages derived from those lineages
will diverge in their turn. A snapshot taken in the
future should allow us to determine, broadly, who has
diverged most recently, and which groups have barely
diverged at all, though our ability to do this will itself
depend on the rate of mutation in the DNA region
concerned: too slow and individuals will tend all to
look the same; too fast and each individual sampled
will tend to be so unique that its relationships to
others will be hard to discern. Molecular markers are
therefore chosen, ideally, such that the mutation rate
matches the question being addressed. A study of
differentiation between gerbils living in different
burrow systems in the same, local population should
use a region of DNA where the mutation rate is high
(much divergence from generation to generation);
whereas a study tracing the routes of colonization that
have placed different populations of brown bears
Molecular markers
9781405156585_4_008.qxd 11/5/07 14:54 Page 254

Chapter 8 Evolutionary ecology
255
over the whole of Europe in the 10,000–12,000 years
since the last glaciation should use a region where
the mutation rate is relatively low.
Polymerase chain reaction (PCR)
As a practical point, most studies in molecular eco-
logy, having extracted the DNA from the organism
concerned, use the polymerase chain reaction (PCR)
to amplify the amount of target material such there is
sufficient available for analysis. By therefore being able
to make use of small samples, this has revolutionized
our ability to sample individuals ‘non-invasively’, using
blood, hair, feces or wing clips. Very simply, PCR
requires ‘primers’ that flank the particular sequence
of DNA that is to be amplified. In the PCR reaction,
nowadays fully automated, the originally double-
stranded DNA is denatured to single strands, the
primers anneal to the separated strands, and an
enzyme, DNA polymerase, copies the sequence
between the primers. This series of reactions is then
repeated 30–40 times, and, since the process of
repeated amplification is exponential, an originally
small amount of target DNA in the midst of other,
unwanted sequences becomes a large enough amount
of target to be subjected to analysis. Note, though,
that hidden within this brief description is the need to
have identified not only informative target regions of
DNA, but also the primers that characterize them.
Nuclear and mitochondrial DNA
In the past especially, many studies have used not
nuclear DNA (inherited equally from both parents and
holding the code for the vast majority of an organism’s
functions) but the relatively small lengths of mito-
chondrial DNA (mtDNA), found in the mitochondria in
the cytoplasm of each of an organism’s cells. The
main advantages of mtDNA are that, almost always, it
is inherited only from the mother (who contributes the
cytoplasm to the fused egg) and does not undergo
recombination. Thus, lineages can be more clearly
traced from generation to generation. Also, the muta-
tion rate is higher than for coding regions of nuclear
DNA, allowing finer resolution differentiation. On the
other hand, mtDNA offers only a small number of
targets, and its maternal inheritance means that when
disparate types meet in a population it is impossible
to know whether any individuals are the result of
matings between them. Increasingly, therefore, studies
are focusing on regions of nuclear DNA, though often
in parallel with analyses of mtDNA genes, combining
the advantages of both.
Microsatellites
Within the nuclear genome, sequences coding for
proteins (i.e. genes) are by no means the only regions
that have been utilized by molecular biologists. Micro-
satellites, for example, are regions of DNA in which
the same two, three of four bases are repeated many
times, preceded and followed in the sequence by
flanking regions that uniquely identify each micro-
satellite (Figure 8.2a). The variability comes from the
fact that the number of ‘repeats’ can vary, the result-
ing lengths of microsatellite DNA being measured by
the speed at which they move through a semisolid
medium (a ‘gel’) under the influence of an electric
current (electrophoresis). Microsatellites may be highly
polymorphic within a population. Thus, an appropriately
chosen ‘panel’ of microsatellites for a species may
effectively allow each individual in a population to be
uniquely identified (a DNA ‘fingerprint’), making micro-
satellites especially appropriate at the finer scales of
differentiation.
Sequencing
As far as nuclear or mitochondrial genes are con-
cerned, having chosen, extracted and amplified
the target region from a sample of individuals, it is
necessary to have some basis for differentiating indi-
viduals from one another, determining who is most
similar to whom, and so on. Increasingly, as automa-
tion improves, and costs come down, the whole
sequences of genes are being determined. As previ-
ously noted, regions of the same gene differ in terms
of their functional importance (Figure 8.2b). Some
regions are ‘conserved’ from individual to individual,
from population to population, and often from species
to species. These are (or are presumed to be) the
regions of greatest functional importance, and they
play effectively no part in differentiation. But there are
other regions where far more variation is observed
(and that can be presumed, therefore, to be neutral
or at least subject to weaker selective constraints),
and it is on the basis of this that individuals and
populations can be differentiated.
s
9781405156585_4_008.qxd 11/5/07 14:54 Page 255

8.2.1 Differentiation within species
Albatrosses, wide ranging sea birds with the largest wingspans of any birds alive
today, have achieved iconic status by virtue of their appearance in poems and
stories, but of the 21 species normally recognized, 19 are regarded as ‘threatened’
with extinction and the other two as ‘near threatened’. The black-browed
albatross has recently been split by taxonomists into two species: Thalassarche
impavida, found only on Campbell Island, between New Zealand and Antarctica,
and T. melanophris, with breeding populations elsewhere in the sub-Antarctic,
including the Falkland Islands, South Georgia and Chile (Figure 8.3a). The
gray-headed albatross, T. chrysostoma, similar in size, also breeds on a number of
sub-Antarctic islands, including South Georgia. The black-browed species usu-
ally remain associated with coastal shelf systems, whereas gray-headed albatrosses
are far more ‘oceanic’ in their feeding grounds, but both, like all albatross species,
are thought to return very close to their place of birth to breed (natal philopatry).
Part III Individuals, Populations, Communities and Ecosystems
256
Restriction fragment length polymorphism
(RFLP)
However, in the past especially, use was often made
of ‘restriction endonuclease’ enzymes that cut DNA
at specific recognition sites situated along its length
and so split an original strand of DNA into fragments.
Individuals differ, as a result of largely neutral muta-
tions, in the location of these sites, and so they
differ, too, in the lengths of the fragments generated,
these lengths being monitored by electrophoresis.
This variation, within a population, is known as
restriction fragment length polymorphism, RFLP,
and there are therefore separate polymorphisms for
different restriction enzymes (because their recogni-
tion sites differ). Samples can thus be subjected
in turn to a series of enzymes, and the most differ-
entiated individuals will then differ in the greatest
number of RFLPs. Its disadvantage, of course, is that
it utilizes only a small part of the underlying sequence
variation.
s
Allele 1 which has 10 repeats
(b)
Allele 2 which has 8 repeats
Individual 1
Individual 2
Individual 3
Individual 4
Individual 5
(a)
Flanking region Flanking regionMicrosatellite
Figure 8.2
(a) A ‘locus’, here, refers to the location of a region in
the overall DNA sequence. An ‘allele’ is the particular
variant of sequence that exists at that locus in a
particular case. Remember that that sequence is of
two strands of DNA, between which the bases are
paired: G with C and A with T. This figure shows two
contrasting alleles at a microsatellite locus, with its
sequence of repeated bases (of differing length)
in the two DNA strands (red) and exactly similar
flanking regions at either end (black). (b) This figure,
by contrast, shows the base sequence in just one
DNA strand of a hypothetical gene (i.e. a sequence of
DNA coding for a protein) from five individuals. Note
the contrast between the conserved (unvarying)
regions at either end, in black, and a variable region
in red towards the center. Differentiation between
individuals clearly depends on this variable region.
albatrosses
9781405156585_4_008.qxd 11/5/07 14:54 Page 256
Chapter 8 Evolutionary ecology
257
(a)
(b)
(c)
T. impavida
T. melanophris
(Falkland)
T. melanophris
(Diego/South Georgia/Kerguelen)
DR
FI
FI
FI
FI
FI
FI
FI
FI
FI
FI
FI
FI
FI
E
F
mC
mC
mC
mC
mC
mC
mC
mC
A
A
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
BI
B
D
K
K
DR
DR
DR
DR
DR
DR
DR
DR
DR
DR
DR
DR
C
C
C
C
C
C
C
H
G
iC
iC
iC
iC
iC
iC
iC
iC
iC
iC
iC
iC
iC
K
K
K
K
K
K
K
K
K
M
M
M
M
M
M
M
M
M
M
M
B
Campbell Is.
Kerguelen Is.
Marion Is.
Falkland Is.
South Georgia
Diego Ramirez
Antarctica
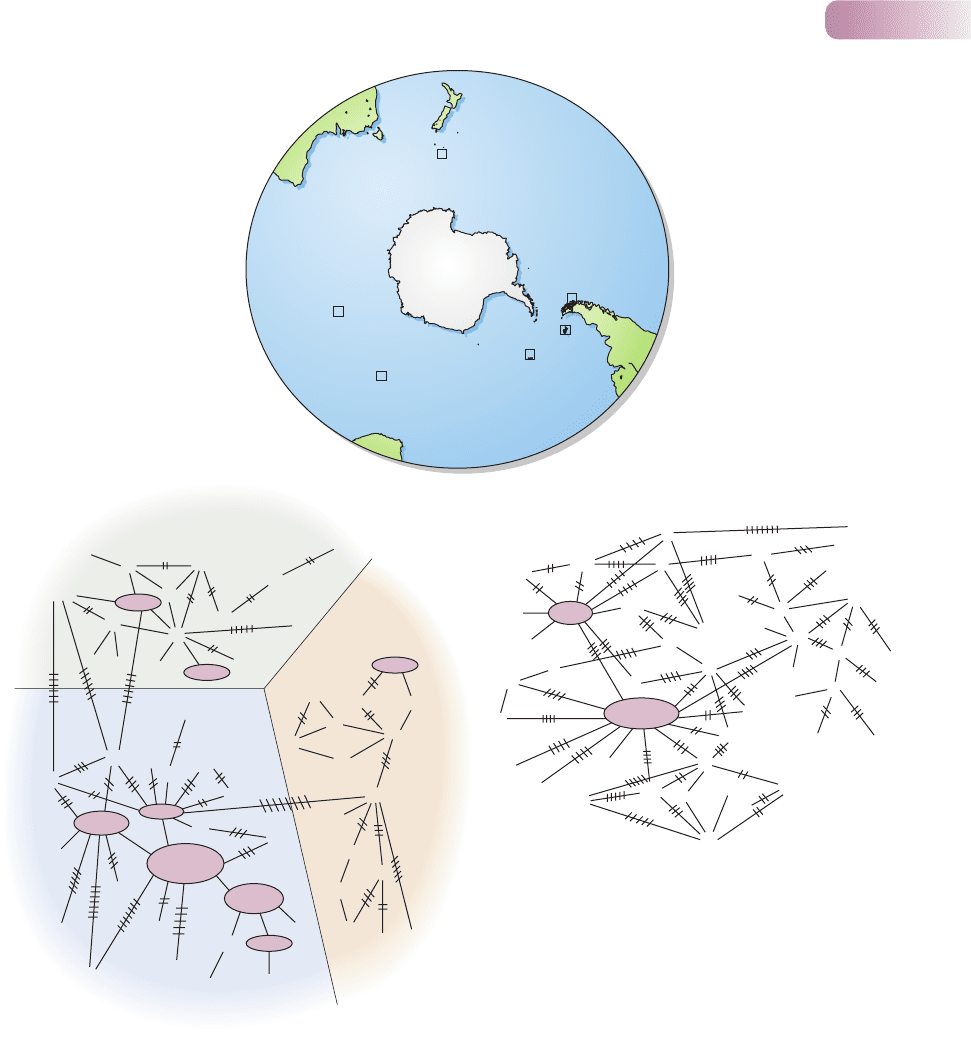
Figure 8.3
Population differentiation in albatrosses: black-browed albatrosses, Thalassarche melanophris and T. impavida, and the gray-headed albatross,
T. chrysostoma. (a) Distribution of sites in the sub-Antarctic from which samples were taken. (b) The relationships amongst 73 black-browed
albatrosses in the base sequence at a focal, variable site in their mtDNA. Where individuals from the same site shared exactly the same sequence,
those individuals have been assigned a letter code (A, B, etc.) and placed in an oval proportional in size to the number of individuals. Individuals that
do not fall into these groups, having a sequence unique within the data set, are identified as follows: BI, South Georgia, DR, Diego Ramirez (Chile),
FI, Falkland Islands, K, Kerguelen Island (all T. melanophris); mC, T. melanophris from Campbell Island; and iC, T. impavida from Campbell Island.
The cross-hatches represent the number of base differences between the individuals (or groups) they join. The samples fall into three ‘clusters’:
T. impavida, T. melanophris from the Falkland Islands and T. melanophris from all other sites. Note though that the clustering is not perfect – as is
normal, like the separation between the populations – and that some of the T. melanophris found on Campbell Island were identifiable as T.
melanophris–T. impavida hybrids. (c) The relationships amongst 50 gray-headed albatrosses in the base sequence at a focal, variable site in their
mtDNA. Coding is the same as in (b) except that M is Marion Island and C is Campbell Island. No separate clusters are discernable in this case.
AFTER BURG & CROXALL, 2001
9781405156585_4_008.qxd 11/5/07 14:54 Page 257
With numbers in all sites declining year on year, therefore, the questions arise:
‘How connected or separate are these populations? Should conservation efforts
be directed at what are currently perceived to be whole species, or at particular
breeding populations?’
These questions were addressed, in both species, by a study that used both
mtDNA sequences and a panel of seven microsatellites (Burg & Croxall, 2001).
The results were clearest for mtDNA (Figure 8.3b, c), but those for the micro-
satellites told the same story. For the black-browed species (Figure 8.3b), the
molecular data confirmed the taxonomists’ view that T. impavida was a separate
species, but also demonstrated breeding between this species and T. melanophris
on Campbell Island and indeed the production of hybrids between these two
species there. More surprisingly, these data also demonstrated that the Falkland
Islands support a breeding population of T. melanophris that is quite separate
from an effectively indivisible population shared by Diego Ramirez (Chile), South
Georgia and Kerguelen Island (in spite of the natal philopatry to these three sites).
By contrast, the wider ranging gray-headed albatrosses, from all five of their sites,
seemed to represent a single breeding population (Figure 8.3c) – again in spite of
their natal philopatry.
From a conservation point of view, though, the most important conclusion
relates to T. melanophris. Whereas previously the relative stability of the large
Falkland Islands population was taken as insurance against a real vulnerability of
the species to extinction, now, in the light of these molecular data, the Falkland
Islands population should be considered as somewhat separate from the rest of
the species, which itself is far more threatened with extinction than was previously
appreciated. (A more active and immediate role for molecular markers in practical
matters of conservation is described in Box 8.2.)
Part III Individuals, Populations, Communities and Ecosystems
258
8.2 TOPICAL ECONCERNS
8.2 Topical ECOncerns
As we shall discuss more fully in Chapter 12, there
is an increasingly frequent conflict between exploit-
ing natural populations as a necessary source of
food and conserving those same populations, both
as an end in itself and so that future generations have
something to eat. In Canada, for example, Pacific
salmonid fish are harvested from a large number
of commercial (industrial) and sport fisheries, each
managed in its own way in an attempt to ensure its
continued viability. So, for instance, a fishery may
be closed altogether at times when fish from other
sources are readily available, in order to allow the
stock to breed and recover. Nonetheless, threats to
sustainability are very real: 2002 saw the first designa-
tion of a Canadian salmon stock, the Interior Fraser
River coho salmon, as ‘endangered’, and many others
require careful protection.
In an ideal world, policing, and hence management,
of the different fisheries would be perfectly effective.
But in reality, illegal fishing is bound to take place and
cannot necessarily be countered simply by catching
offenders ‘in the act’. An alternative, then, or at least
another weapon in the managers’ armoury, is to be
able to identify fish as having been illegally obtained
at some other point in the chain from being caught to
being eaten. Molecular markers make this possible.
The forensic analysis of the origins of our food
molecular markers in
conservation
9781405156585_4_008.qxd 11/5/07 14:54 Page 258
